Genetic and Genomic Resources for Soybean Breeding Research
- PMID: 35567182
- PMCID: PMC9101001
- DOI: 10.3390/plants11091181
Genetic and Genomic Resources for Soybean Breeding Research
Abstract
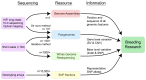
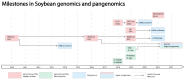
Soybean (Glycine max) is a legume species of significant economic and nutritional value. The yield of soybean continues to increase with the breeding of improved varieties, and this is likely to continue with the application of advanced genetic and genomic approaches for breeding. Genome technologies continue to advance rapidly, with an increasing number of high-quality genome assemblies becoming available. With accumulating data from marker arrays and whole-genome resequencing, studying variations between individuals and populations is becoming increasingly accessible. Furthermore, the recent development of soybean pangenomes has highlighted the significant structural variation between individuals, together with knowledge of what has been selected for or lost during domestication and breeding, information that can be applied for the breeding of improved cultivars. Because of this, resources such as genome assemblies, SNP datasets, pangenomes and associated databases are becoming increasingly important for research underlying soybean crop improvement.
Keywords: assemblies; breeding; databases; genetic variation; genetics; genomics; germplasm; pangenome; soybean.
Conflict of interest statement
The authors declare no conflict of interests.
Figures
Similar articles
-
Legume Pangenome: Status and Scope for Crop Improvement.Plants (Basel). 2022 Nov 10;11(22):3041. doi: 10.3390/plants11223041. Plants (Basel). 2022. PMID: 36432770 Free PMC article. Review.
-
Pangenomes as a Resource to Accelerate Breeding of Under-Utilised Crop Species.Int J Mol Sci. 2022 Feb 28;23(5):2671. doi: 10.3390/ijms23052671. Int J Mol Sci. 2022. PMID: 35269811 Free PMC article. Review.
-
Soybean (Glycine max) Mutant and Germplasm Resources: Current Status and Future Prospects.Curr Protoc Plant Biol. 2016 Mar;1(2):307-327. doi: 10.1002/cppb.20015. Curr Protoc Plant Biol. 2016. PMID: 30775866
-
Construction and comparison of three reference-quality genome assemblies for soybean.Plant J. 2019 Dec;100(5):1066-1082. doi: 10.1111/tpj.14500. Epub 2019 Oct 28. Plant J. 2019. PMID: 31433882
-
The elite variations in germplasms for soybean breeding.Mol Breed. 2023 May 2;43(5):37. doi: 10.1007/s11032-023-01378-0. eCollection 2023 May. Mol Breed. 2023. PMID: 37312749 Free PMC article.
Cited by
-
QTL Mapping and Data Mining to Identify Genes Associated with Soybean Epicotyl Length Using Cultivated Soybean and Wild Soybean.Int J Mol Sci. 2024 Mar 14;25(6):3296. doi: 10.3390/ijms25063296. Int J Mol Sci. 2024. PMID: 38542270 Free PMC article.
-
Exploiting genetic and genomic resources to enhance productivity and abiotic stress adaptation of underutilized pulses.Front Genet. 2023 Jun 16;14:1193780. doi: 10.3389/fgene.2023.1193780. eCollection 2023. Front Genet. 2023. PMID: 37396035 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

