Acridine Based N-Acylhydrazone Derivatives as Potential Anticancer Agents: Synthesis, Characterization and ctDNA/HSA Spectroscopic Binding Properties
- PMID: 35566236
- PMCID: PMC9100673
- DOI: 10.3390/molecules27092883
Acridine Based N-Acylhydrazone Derivatives as Potential Anticancer Agents: Synthesis, Characterization and ctDNA/HSA Spectroscopic Binding Properties
Abstract
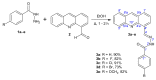
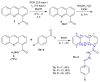
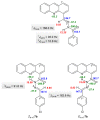
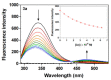
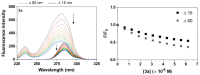
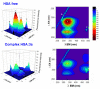
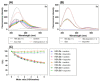
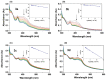
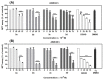
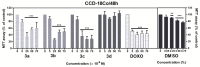
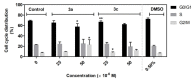
A series of novel acridine N-acylhydrazone derivatives have been synthesized as potential topoisomerase I/II inhibitors, and their binding (calf thymus DNA—ctDNA and human serum albumin—HSA) and biological activities as potential anticancer agents on proliferation of A549 and CCD-18Co have been evaluated. The acridine-DNA complex 3b (-F) displayed the highest Kb value (Kb = 3.18 × 103 M−1). The HSA-derivatives interactions were studied by fluorescence quenching spectra. This method was used for the calculation of characteristic binding parameters. In the presence of warfarin, the binding constant values were found to decrease (KSV = 2.26 M−1, Kb = 2.54 M−1), suggesting that derivative 3a could bind to HSA at Sudlow site I. The effect of tested derivatives on metabolic activity of A549 cells evaluated by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide or MTT assay decreased as follows 3b(-F) > 3a(-H) > 3c(-Cl) > 3d(-Br). The derivatives 3c and 3d in vitro act as potential dual inhibitors of hTopo I and II with a partial effect on the metabolic activity of cancer cells A594. The acridine-benzohydrazides 3a and 3c reduced the clonogenic ability of A549 cells by 72% or 74%, respectively. The general results of the study suggest that the novel compounds show potential for future development as anticancer agents.
Keywords: HSA binding; acridine; benzohydrazide; ctDNA; spectroscopic study.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Correlation between DNA/HSA-interactions and antimalarial activity of acridine derivatives: Proposing a possible mechanism of action.J Photochem Photobiol B. 2018 Dec;189:165-175. doi: 10.1016/j.jphotobiol.2018.10.016. Epub 2018 Oct 19. J Photochem Photobiol B. 2018. PMID: 30366283
-
Fluorinated 3,6,9-trisubstituted acridine derivatives as DNA interacting agents and topoisomerase inhibitors with A549 antiproliferative activity.Bioorg Chem. 2020 Jan;94:103393. doi: 10.1016/j.bioorg.2019.103393. Epub 2019 Oct 23. Bioorg Chem. 2020. PMID: 31679839
-
Binding site identification of anticancer drug gefitinib to HSA and DNA in the presence of five different probes.J Biomol Struct Dyn. 2019 Mar;37(4):823-836. doi: 10.1080/07391102.2018.1441073. Epub 2018 Feb 22. J Biomol Struct Dyn. 2019. PMID: 29447084
-
A new look at 9-substituted acridines with various biological activities.J Appl Toxicol. 2021 Jan;41(1):175-189. doi: 10.1002/jat.4072. Epub 2020 Sep 24. J Appl Toxicol. 2021. PMID: 32969520 Review.
-
DNA-binding antitumor agents: from pyrimido[5,6,1-de]acridines to other intriguing classes of acridine derivatives.Curr Med Chem. 2002 Sep;9(18):1701-16. doi: 10.2174/0929867023369268. Curr Med Chem. 2002. PMID: 12171552 Review.
Cited by
-
BiCl3-catalyzed green synthesis of 4-hydroxy-2-quinolone analogues under microwave irradiation.RSC Adv. 2023 Sep 21;13(40):28030-28041. doi: 10.1039/d3ra05289c. eCollection 2023 Sep 18. RSC Adv. 2023. PMID: 37746335 Free PMC article.
-
In vitro study of pinostrobin propionate and pinostrobin butyrate: Cytotoxic activity against breast cancer cell T47D and its selectivity index.J Public Health Afr. 2023 Mar 16;14(Suppl 1):2516. doi: 10.4081/jphia.2023.2516. eCollection 2023 Mar 30. J Public Health Afr. 2023. PMID: 37492547 Free PMC article.
-
Derivatives Incorporating Acridine, Pyrrole, and Thiazolidine Rings as Promising Antitumor Agents.Molecules. 2023 Sep 14;28(18):6616. doi: 10.3390/molecules28186616. Molecules. 2023. PMID: 37764394 Free PMC article.
-
A novel dihydroacridine derivative targets epidermal growth factor receptor-expressing cancer cells in vitro and in vivo.J Adv Pharm Technol Res. 2024 Apr-Jun;15(2):104-110. doi: 10.4103/JAPTR.JAPTR_392_23. Epub 2024 May 6. J Adv Pharm Technol Res. 2024. PMID: 38903549 Free PMC article.
-
Acylhydrazones and Their Biological Activity: A Review.Molecules. 2022 Dec 9;27(24):8719. doi: 10.3390/molecules27248719. Molecules. 2022. PMID: 36557851 Free PMC article. Review.
References
-
- Dorababu A. Coumarin-Heterocycle Framework: A Privileged Approach in Promising Anticancer Drug Design. Eur. J. Med. Chem. Rep. 2021;2:100006. doi: 10.1016/j.ejmcr.2021.100006. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

