Characteristics of microRNAs and Target Genes in Maize Root under Drought Stress
- PMID: 35563360
- PMCID: PMC9104622
- DOI: 10.3390/ijms23094968
Characteristics of microRNAs and Target Genes in Maize Root under Drought Stress
Abstract
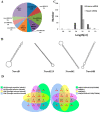
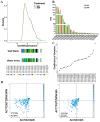
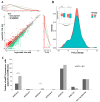
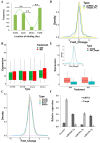
Maize (Zea mays) is an important multi-functional crop. The growth and yield of maize are severely affected by drought stress. Previous studies have shown that microRNAs (miRNAs) in maize play important roles in response to abiotic stress; however, their roles in response to drought stress in maize roots is unclear. In our study, we found 375 miRNAs in the roots of 16 inbred lines. Of the 16 lines, zma-MIR168, zma-MIR156, and zma-MIR166 were highly expressed, whereas zma-MIR399, zma-MIR2218, and zma-MIR2275 exhibited low expression levels. The expression patterns of miRNA in parental lines and their derived RILs are different. Over 50% of miRNAs exhibited a lower expression in recombinant inbred lines than in parents. The expression of 50 miRNAs was significantly altered under water stress (WS) in at least three inbred lines, and the expression of miRNAs in drought-tolerant lines changed markedly. To better understand the reasons for miRNA response to drought, the degree of histone modifications for miRNA genes was estimated. The methylation level of H3K4 and H3K9 in miRNA precursor regions changed more noticeably after WS, but no such phenomenon was seen for DNA methylation and m6A modification. After the prediction of miRNA targets using psRNATarget and psRobot, we used correlation analysis and qRT-PCR to further investigate the relationship between miRNAs and target genes. We found that 87 miRNA-target pairs were significantly negatively correlated. In addition, a weighted gene co-expression network analysis using miRNAs, as well as their predicted targets, was conducted to reveal that miR159, miR394, and miR319 may be related to maize root growth. The results demonstrated that miRNAs might play essential roles in the response to drought stress.
Keywords: drought stress; maize; miRNA; root; target genes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Maize miRNAs and their putative target genes involved in chilling stress response in 5-day old seedlings.BMC Genomics. 2024 May 15;25(1):479. doi: 10.1186/s12864-024-10403-1. BMC Genomics. 2024. PMID: 38750515 Free PMC article.
-
Transcriptional regulatory networks in response to drought stress and rewatering in maize (Zea mays L.).Mol Genet Genomics. 2021 Nov;296(6):1203-1219. doi: 10.1007/s00438-021-01820-y. Epub 2021 Oct 3. Mol Genet Genomics. 2021. PMID: 34601650 Review.
-
Identification and Characterization of Novel Maize Mirnas Involved in Different Genetic Background.Int J Biol Sci. 2015 May 22;11(7):781-93. doi: 10.7150/ijbs.11619. eCollection 2015. Int J Biol Sci. 2015. PMID: 26078720 Free PMC article.
-
Expression of zma-miR169 miRNAs and their target ZmNF-YA genes in response to abiotic stress in maize leaves.Gene. 2015 Jan 25;555(2):178-85. doi: 10.1016/j.gene.2014.11.001. Epub 2014 Nov 5. Gene. 2015. PMID: 25445264
-
Transcriptome expression profiles reveal response mechanisms to drought and drought-stress mitigation mechanisms by exogenous glycine betaine in maize.Biotechnol Lett. 2022 Mar;44(3):367-386. doi: 10.1007/s10529-022-03221-6. Epub 2022 Mar 16. Biotechnol Lett. 2022. PMID: 35294695 Review.
Cited by
-
Identification and Functional Analysis of Drought-Responsive Long Noncoding RNAs in Maize Roots.Int J Mol Sci. 2023 Oct 10;24(20):15039. doi: 10.3390/ijms242015039. Int J Mol Sci. 2023. PMID: 37894720 Free PMC article.
-
Genetic, molecular and physiological crosstalk during drought tolerance in maize (Zea mays): pathways to resilient agriculture.Planta. 2024 Aug 28;260(4):81. doi: 10.1007/s00425-024-04517-9. Planta. 2024. PMID: 39196449 Review.
-
Comparative Genome-Wide Analysis of MicroRNAs and Their Target Genes in Roots of Contrasting Indica Rice Cultivars under Reproductive-Stage Drought.Genes (Basel). 2023 Jul 1;14(7):1390. doi: 10.3390/genes14071390. Genes (Basel). 2023. PMID: 37510295 Free PMC article.
-
MicroRNA (miRNA) profiling of maize genotypes with differential response to Aspergillus flavus implies zma-miR156-squamosa promoter binding protein (SBP) and zma-miR398/zma-miR394-F -box combinations involved in resistance mechanisms.Stress Biol. 2024 May 10;4(1):26. doi: 10.1007/s44154-024-00158-w. Stress Biol. 2024. PMID: 38727957 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

