Construction and application of star polycation nanocarrier-based microRNA delivery system in Arabidopsis and maize
- PMID: 35525952
- PMCID: PMC9077854
- DOI: 10.1186/s12951-022-01443-4
Construction and application of star polycation nanocarrier-based microRNA delivery system in Arabidopsis and maize
Abstract
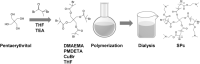
Background: MicroRNA (miRNA) plays vital roles in the regulation of both plant architecture and stress resistance through cleavage or translation inhibition of the target messenger RNAs (mRNAs). However, miRNA-induced gene silencing remains a major challenge in vivo due to the low delivery efficiency and instability of miRNA, thus an efficient and simple method is urgently needed for miRNA transformation. Previous researches have constructed a star polycation (SPc)-mediated transdermal double-stranded RNA (dsRNA) delivery system, achieving efficient dsRNA delivery and gene silencing in insect pests.
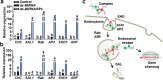
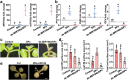
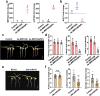
Results: Here, we tested SPc-based platform for direct delivery of double-stranded precursor miRNA (ds-MIRNA) into protoplasts and plants. The results showed that SPc could assemble with ds-MIRNA through electrostatic interaction to form nano-sized ds-MIRNA/SPc complex. The complex could penetrate the root cortex and be systematically transported through the vascular tissue in seedlings of Arabidopsis and maize. Meanwhile, the complex could up-regulate the expression of endocytosis-related genes in both protoplasts and plants to promote the cellular uptake. Furthermore, the SPc-delivered ds-MIRNA could efficiently increase mature miRNA amount to suppress the target gene expression, and the similar phenotypes of Arabidopsis and maize were observed compared to the transgenic plants overexpressing miRNA.
Conclusion: To our knowledge, we report the first construction and application of star polycation nanocarrier-based platform for miRNA delivery in plants, which explores a new enable approach of plant biotechnology with efficient transformation for agricultural application.
Keywords: Gene silencing; MiRNA delivery; Nanoparticle; Plant biotechnology; Star polycation.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing financial interest.
Figures
Similar articles
-
Visualization of the process of a nanocarrier-mediated gene delivery: stabilization, endocytosis and endosomal escape of genes for intracellular spreading.J Nanobiotechnology. 2022 Mar 9;20(1):124. doi: 10.1186/s12951-022-01336-6. J Nanobiotechnology. 2022. PMID: 35264206 Free PMC article.
-
Identification of miRNAs and their target genes in developing maize ears by combined small RNA and degradome sequencing.BMC Genomics. 2014 Jan 14;15:25. doi: 10.1186/1471-2164-15-25. BMC Genomics. 2014. PMID: 24422852 Free PMC article.
-
Widespread occurrence of microRNA-mediated target cleavage on membrane-bound polysomes.Genome Biol. 2021 Jan 5;22(1):15. doi: 10.1186/s13059-020-02242-6. Genome Biol. 2021. PMID: 33402203 Free PMC article.
-
Signal transduction in maize and Arabidopsis mesophyll protoplasts.Plant Physiol. 2001 Dec;127(4):1466-75. Plant Physiol. 2001. PMID: 11743090 Free PMC article. Review.
-
Progress in microRNA delivery.J Control Release. 2013 Dec 28;172(3):962-74. doi: 10.1016/j.jconrel.2013.09.015. Epub 2013 Sep 25. J Control Release. 2013. PMID: 24075926 Free PMC article. Review.
Cited by
-
Nanomaterial inactivates environmental virus and enhances plant immunity for controlling tobacco mosaic virus disease.Nat Commun. 2024 Oct 1;15(1):8509. doi: 10.1038/s41467-024-52851-z. Nat Commun. 2024. PMID: 39353964 Free PMC article.
-
A Preparation Method of Nano-Pesticide Improves the Selective Toxicity toward Natural Enemies.Nanomaterials (Basel). 2022 Jul 14;12(14):2419. doi: 10.3390/nano12142419. Nanomaterials (Basel). 2022. PMID: 35889640 Free PMC article.
-
Improving RNA-based crop protection through nanotechnology and insights from cross-kingdom RNA trafficking.Curr Opin Plant Biol. 2023 Dec;76:102441. doi: 10.1016/j.pbi.2023.102441. Epub 2023 Sep 9. Curr Opin Plant Biol. 2023. PMID: 37696727 Free PMC article. Review.
-
Calcium nutrition nanoagent rescues tomatoes from mosaic virus disease by accelerating calcium transport and activating antiviral immunity.Front Plant Sci. 2022 Dec 6;13:1092774. doi: 10.3389/fpls.2022.1092774. eCollection 2022. Front Plant Sci. 2022. PMID: 36561462 Free PMC article.
-
Recent Advances in Nanoparticle-Mediated Co-Delivery System: A Promising Strategy in Medical and Agricultural Field.Int J Mol Sci. 2023 Mar 7;24(6):5121. doi: 10.3390/ijms24065121. Int J Mol Sci. 2023. PMID: 36982200 Free PMC article. Review.
References
-
- Gray GD, Basu S, Wickstrom E. Transformed and immortalized cellular uptake of oligodeoxynucleoside phosphorothioates, 3′-Alkylamino oligodeoxynucleotides, 2′-o-methyl oligoribonucleotides, oligodeoxynucleoside methylphosphonates, and peptide nucleic acids. Biochem Pharmacol. 1997;53:1465–1476. doi: 10.1016/S0006-2952(97)82440-9. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

