A Leak-Free Inducible CRISPRi/a System for Gene Functional Studies in Plasmodium falciparum
- PMID: 35510853
- PMCID: PMC9241666
- DOI: 10.1128/spectrum.02782-21
A Leak-Free Inducible CRISPRi/a System for Gene Functional Studies in Plasmodium falciparum
Abstract
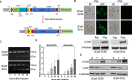
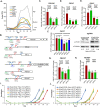
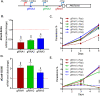
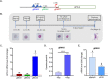
By fusing catalytically dead Cas9 (dCas9) to active domains of histone deacetylase (Sir2a) or acetyltransferase (GCN5), this CRISPR interference/activation (CRISPRi/a) system allows gene regulation at the transcriptional level without causing permanent changes in the parasite genome. However, the constitutive expression of dCas9 poses a challenge for studying essential genes, which may lead to adaptive changes in the parasite, masking the true phenotypes. Here, we developed a leak-free inducible CRISPRi/a system by integrating the DiCre/loxP regulon to allow the expression of dCas9-GCN5/-Sir2a upon transient induction with rapamycin, which allows convenient transcriptional regulation of a gene of interest by introducing a guide RNA targeting its transcription start region. Using eight genes that are either silent or expressed from low to high levels during asexual erythrocytic development, we evaluated the robustness and versatility of this system in the asexual parasites. For most genes analyzed, this inducible CRISPRi/a system led to 1.5- to 3-fold up-or downregulation of the target genes at the mRNA level. Alteration in the expression of PfK13 and PfMYST resulted in altered sensitivities to artemisinin. For autophagy-related protein 18, an essential gene related to artemisinin resistance, a >2-fold up- or downregulation was obtained by inducible CRISPRi/a, leading to growth retardation. For the master regulator of gametocytogenesis, PfAP2-G, a >10-fold increase of the PfAP2-G transcripts was obtained by CRISPRa, resulting in >4-fold higher gametocytemia in the induced parasites. Additionally, inducible CRISPRi/a could also regulate gene expression in gametocytes. This inducible epigenetic regulation system offers a fast way of studying gene functions in Plasmodium falciparum. IMPORTANCE Understanding the fundamental biology of malaria parasites through functional genetic/genomic studies is critical for identifying novel targets for antimalarial development. Conditional knockout/knockdown systems are required to study essential genes in the haploid blood stages of the parasite. In this study, we developed an inducible CRISPRi/a system via the integration of DiCre/loxP. We evaluated the robustness and versatility of this system by activating or repressing eight selected genes and achieved up- and downregulation of the targeted genes located in both the euchromatin and heterochromatin regions. This system offers the malaria research community another tool for functional genetic studies.
Keywords: DiCre; Plasmodium falciparum; dCas9; gene regulation; human malaria.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Investigation of Heterochromatin Protein 1 Function in the Malaria Parasite Plasmodium falciparum Using a Conditional Domain Deletion and Swapping Approach.mSphere. 2021 Feb 3;6(1):e01220-20. doi: 10.1128/mSphere.01220-20. mSphere. 2021. PMID: 33536327 Free PMC article.
-
Role of Plasmodium falciparum Kelch 13 Protein Mutations in P. falciparum Populations from Northeastern Myanmar in Mediating Artemisinin Resistance.mBio. 2020 Feb 25;11(1):e01134-19. doi: 10.1128/mBio.01134-19. mBio. 2020. PMID: 32098812 Free PMC article.
-
Targeted Modulation of Chicken Genes In Vitro Using CRISPRa and CRISPRi Toolkit.Genes (Basel). 2023 Apr 13;14(4):906. doi: 10.3390/genes14040906. Genes (Basel). 2023. PMID: 37107664 Free PMC article.
-
Plasmodium falciparum: multifaceted resistance to artemisinins.Malar J. 2016 Mar 9;15:149. doi: 10.1186/s12936-016-1206-9. Malar J. 2016. PMID: 26955948 Free PMC article. Review.
-
Plasmodium falciparum: epigenetic control of var gene regulation and disease.Subcell Biochem. 2013;61:659-82. doi: 10.1007/978-94-007-4525-4_28. Subcell Biochem. 2013. PMID: 23150271 Review.
Cited by
-
Transforming the CRISPR/dCas9-based gene regulation technique into a forward screening tool in Plasmodium falciparum.iScience. 2024 Mar 27;27(4):109602. doi: 10.1016/j.isci.2024.109602. eCollection 2024 Apr 19. iScience. 2024. PMID: 38617559 Free PMC article.
-
Logical regulation of endogenous gene expression using programmable, multi-input processing CRISPR guide RNAs.Nucleic Acids Res. 2024 Aug 12;52(14):8595-8608. doi: 10.1093/nar/gkae549. Nucleic Acids Res. 2024. PMID: 38943344 Free PMC article.
-
Plasmodium falciparum resistance to artemisinin-based combination therapies.Curr Opin Microbiol. 2022 Oct;69:102193. doi: 10.1016/j.mib.2022.102193. Epub 2022 Aug 22. Curr Opin Microbiol. 2022. PMID: 36007459 Free PMC article. Review.
-
Plasmodium falciparum GCN5 plays a key role in regulating artemisinin resistance-related stress responses.Antimicrob Agents Chemother. 2023 Oct 18;67(10):e0057723. doi: 10.1128/aac.00577-23. Epub 2023 Sep 13. Antimicrob Agents Chemother. 2023. PMID: 37702516 Free PMC article.
-
MYST regulates DNA repair and forms a NuA4-like complex in the malaria parasite Plasmodium falciparum.mSphere. 2024 Apr 23;9(4):e0014024. doi: 10.1128/msphere.00140-24. Epub 2024 Apr 2. mSphere. 2024. PMID: 38564734 Free PMC article.
References
-
- Collins CR, Das S, Wong EH, Andenmatten N, Stallmach R, Hackett F, Herman J-P, Müller S, Meissner M, Blackman MJ. 2013. Robust inducible Cre recombinase activity in the human malaria parasite Plasmodium falciparum enables efficient gene deletion within a single asexual erythrocytic growth cycle. Mol Microbiol 88:687–701. doi:10.1111/mmi.12206. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

