EIF4A3-induced circTOLLIP promotes the progression of hepatocellular carcinoma via the miR-516a-5p/PBX3/EMT pathway
- PMID: 35509064
- PMCID: PMC9069765
- DOI: 10.1186/s13046-022-02378-2
EIF4A3-induced circTOLLIP promotes the progression of hepatocellular carcinoma via the miR-516a-5p/PBX3/EMT pathway
Abstract
Background: Circular RNAs (circRNAs) function as crucial regulators in multiple cancers, including hepatocellular carcinoma (HCC). However, the roles of circRNAs in HCC remains largely unknown.
Methods: circTOLLIP was identified in HCC by screening of two public circRNA microarray datasets and detected in HCC cells and tissues through quantitative real-time PCR (qRT-PCR) and in situ hybridization (ISH). Gain- and loss-of-function assays were performed to confirm the biological effects of circTOLLIP on HCC in vitro and in vivo. Mechanistically, bioinformatics analysis of online databases, MS2-RNA pulldown, biotin-labeled circTOLLIP/miR-516a-5p RNA pulldown, RNA immunoprecipitation (RIP), luciferase reporter assay, fluorescence in situ hybridization assay (FISH) and RNA sequencing were used to confirm the regulation of Eukaryotic initiation factor 4A3 (EIF4A3) on circTOLLIP and the interaction among circTOLLIP, miR-516a-5p and PBX homeobox 3 (PBX3).
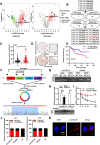
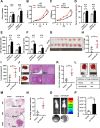
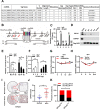
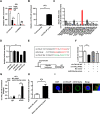
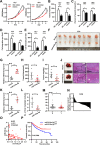
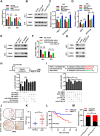
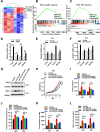
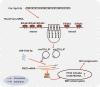
Results: circTOLLIP was significantly upregulated in HCC cells and tissues. High circTOLLIP expression was correlated with poor overall survival (OS) and disease-free survival (DFS) in patients. circTOLLIP promoted the proliferation and metastasis of HCC cells in vitro and in vivo. Mechanistically, EIF4A3 promoted the biogenesis of circTOLLIP without affecting its stability. Moreover, circTOLLIP sponged miR-516a-5p to elevate the expression of PBX3, thereby activating the epithelial-to-mesenchymal transition (EMT) pathway and facilitating tumor progression in HCC.
Conclusions: Our findings indicate that EIF4A3-induced circTOLLIP promotes the progression of HCC through the circTOLLIP/miR-516a-5p/PBX3/EMT axis.
Keywords: EMT; HCC; PBX3; circTOLLIP.
© 2022. The Author(s).
Conflict of interest statement
All authors declare that they have no competing interests.
Figures
Similar articles
-
EIF4A3-regulated circ_0087429 can reverse EMT and inhibit the progression of cervical cancer via miR-5003-3p-dependent upregulation of OGN expression.J Exp Clin Cancer Res. 2022 May 5;41(1):165. doi: 10.1186/s13046-022-02368-4. J Exp Clin Cancer Res. 2022. PMID: 35513835 Free PMC article.
-
Exosome-derived circCCAR1 promotes CD8 + T-cell dysfunction and anti-PD1 resistance in hepatocellular carcinoma.Mol Cancer. 2023 Mar 18;22(1):55. doi: 10.1186/s12943-023-01759-1. Mol Cancer. 2023. PMID: 36932387 Free PMC article.
-
Hsa_circ_0005397 promotes hepatocellular carcinoma progression through EIF4A3.BMC Cancer. 2024 Feb 21;24(1):239. doi: 10.1186/s12885-024-11984-6. BMC Cancer. 2024. PMID: 38383334 Free PMC article.
-
LncRNA SNHG12 promotes proliferation and epithelial mesenchymal transition in hepatocellular carcinoma through targeting HEG1 via miR-516a-5p.Cell Signal. 2021 Aug;84:109992. doi: 10.1016/j.cellsig.2021.109992. Epub 2021 Mar 24. Cell Signal. 2021. PMID: 33774129
-
Circ-CSPP1 knockdown suppresses hepatocellular carcinoma progression through miR-493-5p releasing-mediated HMGB1 downregulation.Cell Signal. 2021 Oct;86:110065. doi: 10.1016/j.cellsig.2021.110065. Epub 2021 Jun 26. Cell Signal. 2021. PMID: 34182091 Review.
Cited by
-
MicroRNAs in Hepatocellular Carcinoma: Insights into Regulatory Mechanisms, Clinical Significance, and Therapeutic Potential.Cancer Manag Res. 2024 Oct 19;16:1491-1507. doi: 10.2147/CMAR.S477698. eCollection 2024. Cancer Manag Res. 2024. PMID: 39450194 Free PMC article. Review.
-
EIF4A3-Induced Circ_0059914 Promoted Angiogenesis and EMT of Glioma via the miR-1249/VEGFA Pathway.Mol Neurobiol. 2025 Jan;62(1):973-987. doi: 10.1007/s12035-024-04319-w. Epub 2024 Jul 1. Mol Neurobiol. 2025. PMID: 38951469
-
The pivotal role of EMT-related noncoding RNAs regulatory axes in hepatocellular carcinoma.Front Pharmacol. 2023 Sep 11;14:1270425. doi: 10.3389/fphar.2023.1270425. eCollection 2023. Front Pharmacol. 2023. PMID: 37767397 Free PMC article. Review.
-
EIF4A3-mediated circ_0042881 activates the RAS pathway via miR-217/SOS1 axis to facilitate breast cancer progression.Cell Death Dis. 2023 Aug 25;14(8):559. doi: 10.1038/s41419-023-06085-4. Cell Death Dis. 2023. PMID: 37626035 Free PMC article.
-
EIF4A3 induced circABCA5 promotes the gastric cancer progression by SPI1 mediated IL6/JAK2/STAT3 signaling.Am J Cancer Res. 2023 Feb 15;13(2):602-622. eCollection 2023. Am J Cancer Res. 2023. PMID: 36895988 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

