Transcriptomic analysis reveals optimal cytokine combinations for SARS-CoV-2-specific T cell therapy products
- PMID: 35506060
- PMCID: PMC9050197
- DOI: 10.1016/j.omtm.2022.04.013
Transcriptomic analysis reveals optimal cytokine combinations for SARS-CoV-2-specific T cell therapy products
Abstract
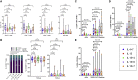
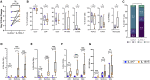
Adoptive T cell immunotherapy has been used to restore immunity against multiple viral targets in immunocompromised patients after bone-marrow transplantation and has been proposed as a strategy for preventing coronavirus 2019 (COVID-19) in this population. Ideally, expanded severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-virus-specific T cells (CSTs) should demonstrate marked cell expansion, T cell specificity, and CD8+ T cell skewing prior to adoptive transfer. However, current methodologies using IL-4 + IL-7 result in suboptimal specificity, especially in CD8+ cells. Using a microexpansion platform, we screened various cytokine cocktails (IL-4 + IL-7, IL-15, IL-15 + IL-4, IL-15 + IL-6, and IL-15 + IL-7) for the most favorable culture conditions. IL-15 + IL-7 optimally balanced T cell expansion, polyfunctionality, and CD8+ T cell skewing of a final therapeutic T cell product. Additionally, the transcriptomes of CD4+ and CD8+ T cells cultured with IL-15 + IL-7 displayed the strongest induction of antiviral type I interferon (IFN) response genes. Subsequently, microexpansion results were successfully translated to a Good Manufacturing Practice (GMP)-applicable format where IL-15 + IL-7 outperformed IL-4 + IL-7 in specificity and expansion, especially in the desirable CD8+ T cell compartment. These results demonstrate the functional implications of IL-15-, IL-4-, and IL-7-containing cocktails for therapeutic T cell expansion, which could have broad implication for cellular therapy, and pioneer the use of RNA sequencing (RNA-seq) to guide viral-specific T cell (VST) product manufacturing.
Keywords: COVID-19; RNA-seq; SARS-CoV-2; cytokines; immunocompromised; manufacturing; therapeutic viral-specific T cells.
© 2022 The Authors.
Conflict of interest statement
P.J.H. is a cofounder and is on the board of directors of Mana Therapeutics. P.J.H. is on a scientific advisory board for Cellevolve. M.D.K. is on a scientific advisory panel for Gilead Sciences. The remaining authors declare no competing financial interests.
Figures

Similar articles
-
Off-the-Shelf Partial HLA Matching SARS-CoV-2 Antigen Specific T Cell Therapy: A New Possibility for COVID-19 Treatment.Front Immunol. 2021 Dec 23;12:751869. doi: 10.3389/fimmu.2021.751869. eCollection 2021. Front Immunol. 2021. PMID: 35003063 Free PMC article.
-
Virus-Specific T Cells From Cryopreserved Blood During an Emergent Virus Outbreak for a Potential Off-the-Shelf Therapy.Transplant Cell Ther. 2023 Sep;29(9):572.e1-572.e13. doi: 10.1016/j.jtct.2023.06.001. Epub 2023 Jun 6. Transplant Cell Ther. 2023. PMID: 37290691
-
Adoptive transfer of ex vivo expanded SARS-CoV-2-specific cytotoxic lymphocytes: A viable strategy for COVID-19 immunosuppressed patients?Transpl Infect Dis. 2021 Aug;23(4):e13602. doi: 10.1111/tid.13602. Epub 2021 Mar 31. Transpl Infect Dis. 2021. PMID: 33728702 Free PMC article.
-
Characterization of a Cytomegalovirus-Specific T Lymphocyte Product Obtained Through a Rapid and Scalable Production Process for Use in Adoptive Immunotherapy.Front Immunol. 2020 Feb 25;11:271. doi: 10.3389/fimmu.2020.00271. eCollection 2020. Front Immunol. 2020. PMID: 32161589 Free PMC article.
-
Disruption of CCR5 signaling to treat COVID-19-associated cytokine storm: Case series of four critically ill patients treated with leronlimab.J Transl Autoimmun. 2021;4:100083. doi: 10.1016/j.jtauto.2021.100083. Epub 2021 Jan 6. J Transl Autoimmun. 2021. PMID: 33521616 Free PMC article. Review.
Cited by
-
Terrific cells for SARS-CoV-2.Haematologica. 2023 Jul 1;108(7):1724-1725. doi: 10.3324/haematol.2022.282273. Haematologica. 2023. PMID: 36475524 Free PMC article. No abstract available.
-
A T cell receptor specific for an HLA-A*03:01-restricted epitope in the endogenous retrovirus ERV-K-Env exhibits limited recognition of its cognate epitope.Mob DNA. 2024 Oct 9;15(1):19. doi: 10.1186/s13100-024-00333-w. Mob DNA. 2024. PMID: 39385229 Free PMC article.
-
Limited Immunogenicity of an HLA-A*03:01-restricted Epitope of Erv-k-env in Non-hiv-1 Settings: Implications for Adoptive Cell Therapy in Cancer.Res Sq [Preprint]. 2024 May 30:rs.3.rs-4432372. doi: 10.21203/rs.3.rs-4432372/v1. Res Sq. 2024. Update in: Mob DNA. 2024 Oct 9;15(1):19. doi: 10.1186/s13100-024-00333-w. PMID: 38854052 Free PMC article. Updated. Preprint.
-
In Silico and In Vitro studies of taiwan chingguan yihau (NRICM101) on TNF-α/IL-1β-induced human lung cells.Biomedicine (Taipei). 2022 Sep 1;12(3):56-71. doi: 10.37796/2211-8039.1378. eCollection 2022. Biomedicine (Taipei). 2022. PMID: 36381194 Free PMC article.
References
-
- Meyts I., Bucciol G., Quinti I., Neven B., Fischer A., Seoane E., Lopez-Granados E., Gianelli C., Robles-Marhuenda A., Jeandel P.Y., et al. Coronavirus Disease 2019 in patients with inborn errors of immunity: an international study. J. Allergy Clin. Immunol. 2021;147:520–531. doi: 10.1016/j.jaci.2020.09.010. - DOI - PMC - PubMed
-
- Vijenthira A., Gong I.Y., Fox T.A., Booth S., Cook G., Fattizzo B., Martin-Moro F., Razanamahery J., Riches J.C., Zwicker J., et al. Outcomes of patients with hematologic malignancies and COVID-19: a systematic review and meta-analysis of 3377 patients. Blood. 2020;136:2881–2892. doi: 10.1182/blood.2020008824. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

