N7-Methylguanosine-Related lncRNAs: Integrated Analysis Associated With Prognosis and Progression in Clear Cell Renal Cell Carcinoma
- PMID: 35495133
- PMCID: PMC9043611
- DOI: 10.3389/fgene.2022.871899
N7-Methylguanosine-Related lncRNAs: Integrated Analysis Associated With Prognosis and Progression in Clear Cell Renal Cell Carcinoma
Abstract
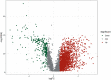
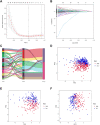
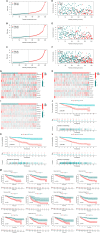
N7-Methylguanosine (m7G) and long non-coding RNAs (lncRNAs) have been widely reported to play an important role in cancer. However, there is little known about the relationship between m7G-related lncRNAs and clear cell renal cell carcinoma (ccRCC). To find new potential biomarkers and construct an m7G-related lncRNA prognostic signature for ccRCC, we retrieved transcriptome data and clinical data from The Cancer Genome Atlas (TCGA), and divided the entire set into train set and test set with the ratio of 1:1 randomly. The m7G-related lncRNAs were identified by Pearson correlation analysis (|coefficients| > 0.4, and p < 0.001). Then we performed the univariate Cox regression and least absolute shrinkage and selection operator (LASSO) Cox regression analysis to construct a 12 m7G-related lncRNA prognostic signature. Next, principal component analysis (PCA), the Kaplan-Meier method, time-dependent receiver operating characteristics (ROC) were made to verify and evaluate the risk signature. A nomogram based on the risk signature and clinical parameters was developed and showed high accuracy and reliability for predicting the overall survival (OS). Functional enrichment analysis (GO, KEGG and GSEA) was used to investigate the potential biological pathways. We also performed the analysis of tumor mutation burden (TMB), immunological analysis including immune scores, immune cell infiltration (ICI), immune function, tumor immune escape (TIE) and immunotherapeutic drug in our study. In conclusion, using the 12 m7G-related lncRNA risk signature as a prognostic indicator may offer us insight into the oncogenesis and treatment response prediction of ccRCC.
Keywords: N7-methylguanosine; clear cell renal cell carcinoma; lncRNA; prognosis; tumor microenvironment; tumor mutation burden.
Copyright © 2022 Ming and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Construction and validation of a prognostic signature for mucinous colonic adenocarcinoma based on N7-methylguanosine-related long non-coding RNAs.J Gastrointest Oncol. 2024 Feb 29;15(1):203-219. doi: 10.21037/jgo-23-980. Epub 2024 Feb 28. J Gastrointest Oncol. 2024. PMID: 38482248 Free PMC article.
-
Construction of prognostic signature of breast cancer based on N7-Methylguanosine-Related LncRNAs and prediction of immune response.Front Genet. 2022 Oct 24;13:991162. doi: 10.3389/fgene.2022.991162. eCollection 2022. Front Genet. 2022. PMID: 36353118 Free PMC article.
-
Predicting prognosis and immune responses in hepatocellular carcinoma based on N7-methylguanosine-related long noncoding RNAs.Front Genet. 2022 Aug 30;13:930446. doi: 10.3389/fgene.2022.930446. eCollection 2022. Front Genet. 2022. PMID: 36110218 Free PMC article.
-
N7-methylguanosine modification: from regulatory roles to therapeutic implications in cancer.Am J Cancer Res. 2023 May 15;13(5):1640-1655. eCollection 2023. Am J Cancer Res. 2023. PMID: 37293166 Free PMC article. Review.
-
M7G-related tumor immunity: novel insights of RNA modification and potential therapeutic targets.Int J Biol Sci. 2024 Jan 27;20(4):1238-1255. doi: 10.7150/ijbs.90382. eCollection 2024. Int J Biol Sci. 2024. PMID: 38385078 Free PMC article. Review.
Cited by
-
Construction and Validation of Protein Expression-related Prognostic Models in Clear Cell Renal Cell Carcinoma.J Cancer. 2023 Mar 27;14(5):793-808. doi: 10.7150/jca.81915. eCollection 2023. J Cancer. 2023. PMID: 37056387 Free PMC article.
-
A novel prognostic N7-methylguanosine-related long non-coding RNA signature in clear cell renal cell carcinoma.Sci Rep. 2023 Oct 27;13(1):18454. doi: 10.1038/s41598-023-45287-w. Sci Rep. 2023. PMID: 37891201 Free PMC article.
-
A Novel m7G-Related Gene Signature Predicts the Prognosis of Colon Cancer.Cancers (Basel). 2022 Nov 10;14(22):5527. doi: 10.3390/cancers14225527. Cancers (Basel). 2022. PMID: 36428620 Free PMC article.
-
Underexplored reciprocity between genome-wide methylation status and long non-coding RNA expression reflected in breast cancer research: potential impacts for the disease management in the framework of 3P medicine.EPMA J. 2023 May 22;14(2):249-273. doi: 10.1007/s13167-023-00323-7. eCollection 2023 Jun. EPMA J. 2023. PMID: 37275549 Free PMC article. Review.
-
A novel prognostic signature based on N7-methylguanosine-related long non-coding RNAs in breast cancer.Front Genet. 2022 Oct 13;13:1030275. doi: 10.3389/fgene.2022.1030275. eCollection 2022. Front Genet. 2022. PMID: 36313442 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

