Caspase-11 interaction with NLRP3 potentiates the noncanonical activation of the NLRP3 inflammasome
- PMID: 35487985
- PMCID: PMC9106893
- DOI: 10.1038/s41590-022-01192-4
Caspase-11 interaction with NLRP3 potentiates the noncanonical activation of the NLRP3 inflammasome
Abstract
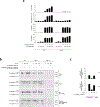
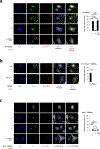
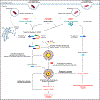
Caspase-11 detection of intracellular lipopolysaccharide (LPS) from invasive Gram-negative bacteria mediates noncanonical activation of the NLRP3 inflammasome. While avirulent bacteria do not invade the cytosol, their presence in tissues necessitates clearance and immune system mobilization. Despite sharing LPS, only live avirulent Gram-negative bacteria activate the NLRP3 inflammasome. Here, we found that bacterial mRNA, which signals bacterial viability, was required alongside LPS for noncanonical activation of the NLRP3 inflammasome in macrophages. Concurrent detection of bacterial RNA by NLRP3 and binding of LPS by pro-caspase-11 mediated a pro-caspase-11-NLRP3 interaction before caspase-11 activation and inflammasome assembly. LPS binding to pro-caspase-11 augmented bacterial mRNA-dependent assembly of the NLRP3 inflammasome, while bacterial viability and an assembled NLRP3 inflammasome were necessary for activation of LPS-bound pro-caspase-11. Thus, the pro-caspase-11-NLRP3 interaction nucleated a scaffold for their interdependent activation explaining their functional reciprocal exclusivity. Our findings inform new vaccine adjuvant combinations and sepsis therapy.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Figures








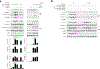





Comment in
-
Dual ligand engagement for noncanonical inflammasome activation.Nat Immunol. 2022 May;23(5):651-653. doi: 10.1038/s41590-022-01188-0. Nat Immunol. 2022. PMID: 35487984 No abstract available.
Similar articles
-
The orphan receptor Nur77 binds cytoplasmic LPS to activate the non-canonical NLRP3 inflammasome.Immunity. 2023 Apr 11;56(4):753-767.e8. doi: 10.1016/j.immuni.2023.03.003. Epub 2023 Mar 30. Immunity. 2023. PMID: 37001519
-
Chalcones Display Anti-NLRP3 Inflammasome Activity in Macrophages through Inhibition of Both Priming and Activation Steps-Structure-Activity-Relationship and Mechanism Studies.Molecules. 2020 Dec 16;25(24):5960. doi: 10.3390/molecules25245960. Molecules. 2020. PMID: 33339319 Free PMC article.
-
The Overexpression of Sirtuin1 (SIRT1) Alleviated Lipopolysaccharide (LPS)-Induced Acute Kidney Injury (AKI) via Inhibiting the Activation of Nucleotide-Binding Oligomerization Domain-Like Receptors (NLR) Family Pyrin Domain Containing 3 (NLRP3) Inflammasome.Med Sci Monit. 2019 Apr 13;25:2718-2726. doi: 10.12659/MSM.913146. Med Sci Monit. 2019. PMID: 30980521 Free PMC article.
-
Structural Mechanisms of NLRP3 Inflammasome Assembly and Activation.Annu Rev Immunol. 2023 Apr 26;41:301-316. doi: 10.1146/annurev-immunol-081022-021207. Epub 2023 Feb 7. Annu Rev Immunol. 2023. PMID: 36750315 Free PMC article. Review.
-
[Advances in mechanisms for NLRP3 inflammasomes regulation].Yao Xue Xue Bao. 2016 Oct;51(10):1505-12. Yao Xue Xue Bao. 2016. PMID: 29924571 Review. Chinese.
Cited by
-
Catalytic activity and autoprocessing of murine caspase-11 mediate noncanonical inflammasome assembly in response to cytosolic LPS.Elife. 2024 Jan 17;13:e83725. doi: 10.7554/eLife.83725. Elife. 2024. PMID: 38231198 Free PMC article.
-
TRIM50 promotes NLRP3 inflammasome activation by directly inducing NLRP3 oligomerization.EMBO Rep. 2022 Nov 7;23(11):e54569. doi: 10.15252/embr.202154569. Epub 2022 Sep 30. EMBO Rep. 2022. PMID: 36178239 Free PMC article.
-
Caspase-5: Structure, Pro-Inflammatory Activity and Evolution.Biomolecules. 2024 Apr 26;14(5):520. doi: 10.3390/biom14050520. Biomolecules. 2024. PMID: 38785927 Free PMC article. Review.
-
Roles of the Caspase-11 Non-Canonical Inflammasome in Rheumatic Diseases.Int J Mol Sci. 2024 Feb 8;25(4):2091. doi: 10.3390/ijms25042091. Int J Mol Sci. 2024. PMID: 38396768 Free PMC article. Review.
-
Exploratory study on the mechanism of necrotic effect of nourishing cells in the context of genital tract infection in premature rupture of membranes.Medicine (Baltimore). 2023 Dec 15;102(50):e36148. doi: 10.1097/MD.0000000000036148. Medicine (Baltimore). 2023. PMID: 38115314 Free PMC article.
References
-
- Kayagaki N et al. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science 341, 1246–1249 (2013). - PubMed
-
- Shi J et al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 514, 187–192 (2014). - PubMed
-
- Broz P & Dixit VM Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol 16, 407–420 (2016). - PubMed
-
- Broz P, Pelegrin P & Shao F The gasdermins, a protein family executing cell death and inflammation. Nat Rev Immunol 20, 143–157 (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

