Impact of Micropolymorphism Outside the Peptide Binding Groove in the Clinically Relevant Allele HLA-C*14 on T Cell Responses in HIV-1 Infection
- PMID: 35475667
- PMCID: PMC9131871
- DOI: 10.1128/jvi.00432-22
Impact of Micropolymorphism Outside the Peptide Binding Groove in the Clinically Relevant Allele HLA-C*14 on T Cell Responses in HIV-1 Infection
Abstract
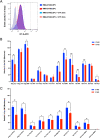
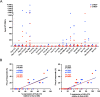
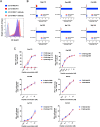
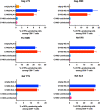
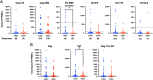
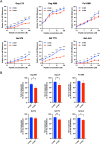
There is increasing evidence for the importance of human leukocyte antigen C (HLA-C)-restricted CD8+ T cells in HIV-1 control, but these responses are relatively poorly investigated. The number of HLA-C-restricted HIV-1 epitopes identified is much smaller than those of HLA-A-restricted or HLA-B-restricted ones. Here, we utilized a mass spectrometry-based approach to identify HIV-1 peptides presented by HLA-C*14:03 protective and HLA-C*14:02 nonprotective alleles. We identified 25 8- to 11-mer HLA-I-bound HIV-1 peptides from HIV-1-infected HLA-C*14:02+/14:03+ cells. Analysis of T cell responses to these peptides identified novel 6 T cell epitopes targeted in HIV-1-infected HLA-C*14:02+/14:03+ subjects. Analyses using HLA stabilization assays demonstrated that all 6 epitope peptides exhibited higher binding to and greater cell surface stabilization of HLA-C*14:02 than HLA-C*14:03. T cell response magnitudes were typically higher in HLA-C*14:02+ than HLA-C*14:03+ individuals, with responses to the Pol KM9 and Nef epitopes being significantly higher. The results show that HLA-C*14:02 can elicit stronger T cell responses to HIV-1 than HLA-C*14:03 and suggest that the single amino acid difference between these HLA-C14 subtypes at position 21, outside the peptide-binding groove, indirectly influences the stability of peptide-HLA-C*14 complexes and induction/expansion of HIV-specific T cells. Taken together with a previous finding that KIR2DL2+ NK cells recognized HLA-C*14:03+ HIV-1-infected cells more than HLA-C*14:02+ ones, the present study indicates that these HLA-C*14 subtypes differentially impact HIV-1 control by T cells and NK cells. IMPORTANCE Some human leukocyte antigen (HLA) class I alleles are associated with good clinical outcomes in HIV-1 infection and are called protective HLA alleles. Identification of T cell epitopes restricted by protective HLA alleles can give important insight into virus-immune system interactions and inform design of immune-based prophylactic/therapeutic strategies. Although epitopes restricted by many protective HLA-A/B alleles have been identified, protective HLA-C alleles are relatively understudied. Here, we identified 6 novel T cell epitopes presented by both HLA-C*14:02 (no association with protection) and HLA-C*14:03 (protective) using a mass spectrometry-based immunopeptidome profiling approach. We found that these peptides bound to and stabilized HLA-C*14:02 better than HLA-C*14:03 and observed differences in induction/expansion of epitope-specific T cell responses in HIV-infected HLA-C*14:02+ versus HLA-C*14:03+ individuals. These results enhance understanding of how the microstructural difference at position 21 between these HLA-C*14 subtypes may influence cellular immune responses involved in viral control in HIV-1 infection.
Keywords: CD8+ T cells; HIV-1; HLA-C; LC-MS/MS; epitopes; peptides.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Identification of Immunodominant HIV-1 Epitopes Presented by HLA-C*12:02, a Protective Allele, Using an Immunopeptidomics Approach.J Virol. 2019 Aug 13;93(17):e00634-19. doi: 10.1128/JVI.00634-19. Print 2019 Sep 1. J Virol. 2019. PMID: 31217245 Free PMC article.
-
Comprehensive screening for human immunodeficiency virus type 1 subtype-specific CD8 cytotoxic T lymphocytes and definition of degenerate epitopes restricted by HLA-A0207 and -C(W)0304 alleles.J Virol. 2002 May;76(10):4971-86. doi: 10.1128/jvi.76.10.4971-4986.2002. J Virol. 2002. PMID: 11967314 Free PMC article.
-
Immunological Control of HIV-1 Disease Progression by Rare Protective HLA Allele.J Virol. 2022 Nov 23;96(22):e0124822. doi: 10.1128/jvi.01248-22. Epub 2022 Nov 3. J Virol. 2022. PMID: 36326273 Free PMC article.
-
T Cell Epitope Discovery in the Context of Distinct and Unique Indigenous HLA Profiles.Front Immunol. 2022 May 6;13:812393. doi: 10.3389/fimmu.2022.812393. eCollection 2022. Front Immunol. 2022. PMID: 35603215 Free PMC article. Review.
-
Viral T-cell epitopes - Identification, characterization and clinical application.Semin Immunol. 2023 Mar;66:101725. doi: 10.1016/j.smim.2023.101725. Epub 2023 Jan 25. Semin Immunol. 2023. PMID: 36706520 Review.
Cited by
-
HIV-1 protective epitope-specific CD8+ T cells in HIV-1-exposed seronegative individuals.iScience. 2023 Sep 29;26(11):108089. doi: 10.1016/j.isci.2023.108089. eCollection 2023 Nov 17. iScience. 2023. PMID: 37867946 Free PMC article.
-
Epitope-dependent effect of long-term cART on maintenance and recovery of HIV-1-specific CD8+ T cells.J Virol. 2023 Nov 30;97(11):e0102423. doi: 10.1128/jvi.01024-23. Epub 2023 Oct 25. J Virol. 2023. PMID: 37877716 Free PMC article.
-
Inclusion body myositis, viral infections, and TDP-43: a narrative review.Clin Exp Med. 2024 May 2;24(1):91. doi: 10.1007/s10238-024-01353-9. Clin Exp Med. 2024. PMID: 38693436 Free PMC article. Review.
References
-
- Altfeld M, Addo MM, Rosenberg ES, Hecht FM, Lee PK, Vogel M, Yu XG, Draenert R, Johnston MN, Strick D, Allen TM, Feeney ME, Kahn JO, Sekaly RP, Levy JA, Rockstroh JK, Goulder PJ, Walker BD. 2003. Influence of HLA-B57 on clinical presentation and viral control during acute HIV-1 infection. AIDS 17:2581–2591. 10.1097/00002030-200312050-00005. - DOI - PubMed
-
- Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, Weale M, Zhang K, Gumbs C, Castagna A, Cossarizza A, Cozzi-Lepri A, De Luca A, Easterbrook P, Francioli P, Mallal S, Martinez-Picado J, Miro JM, Obel N, Smith JP, Wyniger J, Descombes P, Antonarakis SE, Letvin NL, McMichael AJ, Haynes BF, Telenti A, Goldstein DB. 2007. A whole-genome association study of major determinants for host control of HIV-1. Science 317:944–947. 10.1126/science.1143767. - DOI - PMC - PubMed
-
- Lazaryan A, Song W, Lobashevsky E, Tang J, Shrestha S, Zhang K, Gardner LI, McNicholl JM, Wilson CM, Klein RS, Rompalo A, Mayer K, Sobel J, Kaslow RA, Study H, Reaching For Excellence In Adolescent C, Health S, Reaching for Excellence in Adolescent Care and Health Study. 2010. Human leukocyte antigen class I supertypes and HIV-1 control in African Americans. J Virol 84:2610–2617. 10.1128/JVI.01962-09. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

