Reduced IRF4 expression promotes lytic phenotype in Type 2 EBV-infected B cells
- PMID: 35472072
- PMCID: PMC9041801
- DOI: 10.1371/journal.ppat.1010453
Reduced IRF4 expression promotes lytic phenotype in Type 2 EBV-infected B cells
Abstract
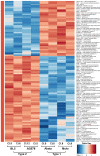
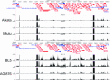
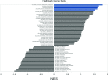
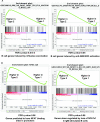
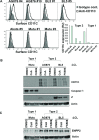
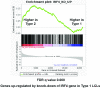
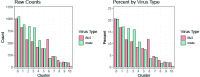
Humans are infected with two types of EBV (Type 1 (T1) and Type 2 (T2)) that differ substantially in their EBNA2 and EBNA 3A/B/C latency proteins and have different phenotypes in B cells. T1 EBV transforms B cells more efficiently than T2 EBV in vitro, and T2 EBV-infected B cells are more lytic. We previously showed that both increased NFATc1/c2 activity, and an NFAT-binding motif within the BZLF1 immediate-early promoter variant (Zp-V3) contained in all T2 strains, contribute to lytic infection in T2 EBV-infected B cells. Here we compare cellular and viral gene expression in early-passage lymphoblastoid cell lines (LCLs) infected with either T1 or T2 EBV strains. Using bulk RNA-seq, we show that T2 LCLs are readily distinguishable from T1 LCLs, with approximately 600 differentially expressed cellular genes. Gene Set Enrichment Analysis (GSEA) suggests that T2 LCLs have increased B-cell receptor (BCR) signaling, NFAT activation, and enhanced expression of epithelial-mesenchymal-transition-associated genes. T2 LCLs also have decreased RNA and protein expression of a cellular gene required for survival of T1 LCLs, IRF4. In addition to its essential role in plasma cell differentiation, IRF4 decreases BCR signaling. Knock-down of IRF4 in a T1 LCL (infected with the Zp-V3-containing Akata strain) induced lytic reactivation whereas over-expression of IRF4 in Burkitt lymphoma cells inhibited both NFATc1 and NFATc2 expression and lytic EBV reactivation. Single-cell RNA-seq confirmed that T2 LCLs have many more lytic cells compared to T1 LCLs and showed that lytically infected cells have both increased NFATc1, and decreased IRF4, compared to latently infected cells. These studies reveal numerous differences in cellular gene expression in B cells infected with T1 versus T2 EBV and suggest that decreased IRF4 contributes to both the latent and lytic phenotypes in cells with T2 EBV.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






Similar articles
-
B cells infected with Type 2 Epstein-Barr virus (EBV) have increased NFATc1/NFATc2 activity and enhanced lytic gene expression in comparison to Type 1 EBV infection.PLoS Pathog. 2020 Feb 14;16(2):e1008365. doi: 10.1371/journal.ppat.1008365. eCollection 2020 Feb. PLoS Pathog. 2020. PMID: 32059024 Free PMC article.
-
IRF4 promotes Epstein-Barr virus activation in Burkitt's lymphoma cells.J Gen Virol. 2019 May;100(5):851-862. doi: 10.1099/jgv.0.001249. Epub 2019 Mar 25. J Gen Virol. 2019. PMID: 30907723
-
A cancer-associated Epstein-Barr virus BZLF1 promoter variant enhances lytic infection.PLoS Pathog. 2018 Jul 27;14(7):e1007179. doi: 10.1371/journal.ppat.1007179. eCollection 2018 Jul. PLoS Pathog. 2018. PMID: 30052684 Free PMC article.
-
Dynamic Epstein-Barr virus gene expression on the path to B-cell transformation.Adv Virus Res. 2014;88:279-313. doi: 10.1016/B978-0-12-800098-4.00006-4. Adv Virus Res. 2014. PMID: 24373315 Free PMC article. Review.
-
Human B-lymphoid cell lines.Hum Cell. 1992 Mar;5(1):25-41. Hum Cell. 1992. PMID: 1329931 Review.
Cited by
-
Functional Targets for Epstein-Barr Virus BART MicroRNAs in B Cell Lymphomas.Cancers (Basel). 2024 Oct 19;16(20):3537. doi: 10.3390/cancers16203537. Cancers (Basel). 2024. PMID: 39456631 Free PMC article.
-
Epstein-Barr virus LMP1 protein promotes proliferation and inhibits differentiation of epithelial cells via activation of YAP and TAZ.Proc Natl Acad Sci U S A. 2023 May 16;120(20):e2219755120. doi: 10.1073/pnas.2219755120. Epub 2023 May 8. Proc Natl Acad Sci U S A. 2023. PMID: 37155846 Free PMC article.
-
A comprehensive single cell data analysis of lymphoblastoid cells reveals the role of super-enhancers in maintaining EBV latency.J Med Virol. 2023 Jan;95(1):e28362. doi: 10.1002/jmv.28362. J Med Virol. 2023. PMID: 36453088 Free PMC article.
-
Shared and distinct interactions of type 1 and type 2 Epstein-Barr Nuclear Antigen 2 with the human genome.BMC Genomics. 2024 Mar 12;25(1):273. doi: 10.1186/s12864-024-10183-8. BMC Genomics. 2024. PMID: 38475709 Free PMC article.
-
Epstein-Barr virus: Biology and clinical disease.Cell. 2022 Sep 29;185(20):3652-3670. doi: 10.1016/j.cell.2022.08.026. Epub 2022 Sep 15. Cell. 2022. PMID: 36113467 Free PMC article. Review.
References
-
- Kieff E, Cohen JI, Longnecker R. Epstein-Barr Virus/Replication and Epstein-Barr Virus. 6th ed. In: Knipe DM, Howley PM, editors. Fields Virology. 6th ed. Philadelphia, PA: Lippincott Williams & Wilkins; 2013. pp. 1898–1959.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

