Sculpting the Bacterial O-Glycoproteome: Functional Analyses of Orthologous Oligosaccharyltransferases with Diverse Targeting Specificities
- PMID: 35471082
- PMCID: PMC9239064
- DOI: 10.1128/mbio.03797-21
Sculpting the Bacterial O-Glycoproteome: Functional Analyses of Orthologous Oligosaccharyltransferases with Diverse Targeting Specificities
Abstract
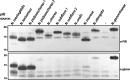
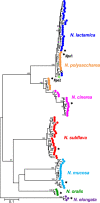
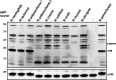
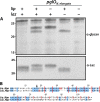
Protein glycosylation systems are widely recognized in bacteria, including members of the genus Neisseria. In most bacterial species, the molecular mechanisms and evolutionary contexts underpinning target protein selection and the glycan repertoire remain poorly understood. Broad-spectrum O-linked protein glycosylation occurs in all human-associated species groups within the genus Neisseria, but knowledge of their individual glycoprotein repertoires is limited. Interestingly, PilE, the pilin subunit of the type IV pilus (Tfp) colonization factor, is glycosylated in Neisseria gonorrhoeae and Neisseria meningitidis but not in the deeply branching species N. elongata subsp. glycolytica. To examine this in more detail, we assessed PilE glycosylation status across the genus and found that PilEs of commensal clade species are not modified by the gonococcal PglO oligosaccharyltransferase. Experiments using PglO oligosaccharyltransferases from across the genus expressed in N. gonorrhoeae showed that although all were capable of broad-spectrum protein glycosylation, those from a deep-branching group of commensals were unable to support resident PilE glycosylation. Further glycoproteomic analyses of these strains using immunoblotting and mass spectrometry revealed other proteins differentially targeted by otherwise remarkably similar oligosaccharyltransferases. Finally, we generated pglO allelic chimeras that begin to localize PglO protein domains associated with unique substrate targeting activities. These findings reveal previously unappreciated differences within the protein glycosylation systems of highly related bacterial species. We propose that the natural diversity manifest in the neisserial protein substrates and oligosaccharyltransferases has significant potential to inform the structure-function relationships operating in these and related bacterial protein glycosylation systems. IMPORTANCE Although general protein glycosylation systems have been well recognized in prokaryotes, the processes governing their distribution, function, and evolution remain poorly understood. Here, we have begun to address these gaps in knowledge by comparative analyses of broad-spectrum O-linked protein glycosylation manifest in species within the genus Neisseria that strictly colonize humans. Using N. gonorrhoeae as a well-defined model organism in conjunction with comparative genomics, intraspecies gene complementation, and glycoprotein phenotyping, we discovered clear differences in both glycosylation susceptibilities and enzymatic targeting activities of otherwise largely conserved proteins. These findings reveal previously unappreciated differences within the protein glycosylation systems of highly related bacterial species. We propose that the natural diversity manifest within Neisseria species has significant potential to elucidate the structure-function relationships operating in these and related systems and to inform novel approaches to applied glycoengineering strategies.
Keywords: Neisseria; evolution; glycoproteins; oligosaccharides; pili.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Characterization of a Unique Tetrasaccharide and Distinct Glycoproteome in the O-Linked Protein Glycosylation System of Neisseria elongata subsp. glycolytica.J Bacteriol. 2015 Oct 19;198(2):256-67. doi: 10.1128/JB.00620-15. Print 2016 Jan 15. J Bacteriol. 2015. PMID: 26483525 Free PMC article.
-
Combining FAIMS based glycoproteomics and DIA proteomics reveals widespread proteome alterations in response to glycosylation occupancy changes in Neisseria gonorrhoeae.Proteomics. 2024 Jul;24(14):e2300496. doi: 10.1002/pmic.202300496. Epub 2024 Feb 15. Proteomics. 2024. PMID: 38361220
-
Disrupted Synthesis of a Di-N-acetylated Sugar Perturbs Mature Glycoform Structure and Microheterogeneity in the O-Linked Protein Glycosylation System of Neisseria elongata subsp. glycolytica.J Bacteriol. 2018 Dec 7;201(1):e00522-18. doi: 10.1128/JB.00522-18. Print 2019 Jan 1. J Bacteriol. 2018. PMID: 30322851 Free PMC article.
-
The role of pilin glycan in neisserial pathogenesis.Mol Cell Biochem. 2003 Nov;253(1-2):179-90. doi: 10.1023/a:1026058311857. Mol Cell Biochem. 2003. PMID: 14619968 Review.
-
Transformation competence and type-4 pilus biogenesis in Neisseria gonorrhoeae--a review.Gene. 1997 Jun 11;192(1):125-34. doi: 10.1016/s0378-1119(97)00038-3. Gene. 1997. PMID: 9224882 Review.
Cited by
-
Glycoproteomic and proteomic analysis of Burkholderia cenocepacia reveals glycosylation events within FliF and MotB are dispensable for motility.Microbiol Spectr. 2024 Jun 4;12(6):e0034624. doi: 10.1128/spectrum.00346-24. Epub 2024 May 6. Microbiol Spectr. 2024. PMID: 38709084 Free PMC article.
-
CRISPRi-Mediated Silencing of Burkholderia O-Linked Glycosylation Systems Enables the Depletion of Glycosylation Yet Results in Modest Proteome Impacts.J Proteome Res. 2023 Jun 2;22(6):1762-1778. doi: 10.1021/acs.jproteome.2c00790. Epub 2023 Mar 30. J Proteome Res. 2023. PMID: 36995114 Free PMC article.
-
Sweet complexity: O-linked protein glycosylation in pathogenic Neisseria.Front Cell Infect Microbiol. 2024 May 14;14:1407863. doi: 10.3389/fcimb.2024.1407863. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38808060 Free PMC article. Review.
References
-
- Iwashkiw JA, Seper A, Weber BS, Scott NE, Vinogradov E, Stratilo C, Reiz B, Cordwell SJ, Whittal R, Schild S, Feldman MF. 2012. Identification of a general O-linked protein glycosylation system in Acinetobacter baumannii and its role in virulence and biofilm formation. PLoS Pathog 8:e1002758. doi:10.1371/journal.ppat.1002758. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
