Paxillin: A Hub for Mechano-Transduction from the β3 Integrin-Talin-Kindlin Axis
- PMID: 35450290
- PMCID: PMC9016114
- DOI: 10.3389/fcell.2022.852016
Paxillin: A Hub for Mechano-Transduction from the β3 Integrin-Talin-Kindlin Axis
Abstract
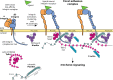
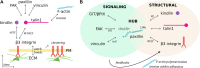
Focal adhesions are specialized integrin-dependent adhesion complexes, which ensure cell anchoring to the extracellular matrix. Focal adhesions also function as mechano-signaling platforms by perceiving and integrating diverse physical and (bio)chemical cues of their microenvironment, and by transducing them into intracellular signaling for the control of cell behavior. The fundamental biological mechanism of creating intracellular signaling in response to changes in tensional forces appears to be tightly linked to paxillin recruitment and binding to focal adhesions. Interestingly, the tension-dependent nature of the paxillin binding to adhesions, combined with its scaffolding function, suggests a major role of this protein in integrating multiple signals from the microenvironment, and accordingly activating diverse molecular responses. This minireview offers an overview of the molecular bases of the mechano-sensitivity and mechano-signaling capacity of core focal adhesion proteins, and highlights the role of paxillin as a key component of the mechano-transducing machinery based on the interaction of cells to substrates activating the β3 integrin-talin1-kindlin.
Keywords: integrin activation; lim domain; mechano-sensing; plasma membrane; tensional force.
Copyright © 2022 Ripamonti, Wehrle-Haller and de Curtis.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Structural and functional analysis of LIM domain-dependent recruitment of paxillin to αvβ3 integrin-positive focal adhesions.Commun Biol. 2021 Mar 29;4(1):380. doi: 10.1038/s42003-021-01886-9. Commun Biol. 2021. PMID: 33782527 Free PMC article.
-
Kindlin supports platelet integrin αIIbβ3 activation by interacting with paxillin.J Cell Sci. 2017 Nov 1;130(21):3764-3775. doi: 10.1242/jcs.205641. Epub 2017 Sep 27. J Cell Sci. 2017. PMID: 28954813 Free PMC article.
-
Structural Basis of Paxillin Recruitment by Kindlin-2 in Regulating Cell Adhesion.Structure. 2019 Nov 5;27(11):1686-1697.e5. doi: 10.1016/j.str.2019.09.006. Epub 2019 Oct 4. Structure. 2019. PMID: 31590942 Free PMC article.
-
Paxillin interactions.J Cell Sci. 2000 Dec;113 Pt 23:4139-40. doi: 10.1242/jcs.113.23.4139. J Cell Sci. 2000. PMID: 11069756 Review.
-
Initiation of focal adhesion assembly by talin and kindlin: A dynamic view.Protein Sci. 2021 Mar;30(3):531-542. doi: 10.1002/pro.4014. Epub 2020 Dec 30. Protein Sci. 2021. PMID: 33336515 Free PMC article. Review.
Cited by
-
Live Cells Imaging and Comparative Phosphoproteomics Uncover Proteins from the Mechanobiome in Entamoeba histolytica.Int J Mol Sci. 2023 May 13;24(10):8726. doi: 10.3390/ijms24108726. Int J Mol Sci. 2023. PMID: 37240072 Free PMC article.
-
Serine/Threonine Protein Phosphatases 1 and 2A in Lung Endothelial Barrier Regulation.Biomedicines. 2023 Jun 5;11(6):1638. doi: 10.3390/biomedicines11061638. Biomedicines. 2023. PMID: 37371733 Free PMC article. Review.
-
Physiological and pathological roles of Hic‑5 in several organs (Review).Int J Mol Med. 2022 Nov;50(5):138. doi: 10.3892/ijmm.2022.5194. Epub 2022 Oct 12. Int J Mol Med. 2022. PMID: 36222304 Free PMC article. Review.
-
Structure, signal transduction, activation, and inhibition of integrin αIIbβ3.Thromb J. 2023 Feb 13;21(1):18. doi: 10.1186/s12959-023-00463-w. Thromb J. 2023. PMID: 36782235 Free PMC article. Review.
-
ConFERMing the role of talin in integrin activation and mechanosignaling.J Cell Sci. 2023 Apr 15;136(8):jcs260576. doi: 10.1242/jcs.260576. Epub 2023 Apr 20. J Cell Sci. 2023. PMID: 37078342 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources

