Dimerization-dependent membrane tethering by Atg23 is essential for yeast autophagy
- PMID: 35443167
- PMCID: PMC9097366
- DOI: 10.1016/j.celrep.2022.110702
Dimerization-dependent membrane tethering by Atg23 is essential for yeast autophagy
Abstract
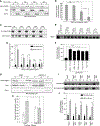
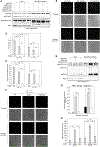
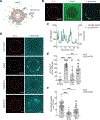
Eukaryotes maintain cellular health through the engulfment and subsequent degradation of intracellular cargo using macroautophagy. The function of Atg23, despite being critical to the efficiency of this process, is unclear due to a lack of biochemical investigations and an absence of any structural information. In this study, we use a combination of in vitro and in vivo methods to show that Atg23 exists primarily as a homodimer, a conformation facilitated by a putative amphipathic helix. We utilize small-angle X-ray scattering to monitor the overall shape of Atg23, revealing that it contains an extended rod-like structure spanning approximately 320 Å. We also demonstrate that Atg23 interacts with membranes directly, primarily through electrostatic interactions, and that these interactions lead to vesicle tethering. Finally, mutation of the hydrophobic face of the putative amphipathic helix completely precludes dimer formation, leading to severely impaired subcellular localization, vesicle tethering, Atg9 binding, and autophagic efficiency.
Keywords: CP: Cell biology; autophagy; lysosome; membrane tether; stress; vacuole.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




Comment in
-
Atg23 is a vesicle-tethering protein.Autophagy. 2022 Oct;18(10):2510-2511. doi: 10.1080/15548627.2022.2105107. Epub 2022 Aug 1. Autophagy. 2022. PMID: 35867625 Free PMC article.
Similar articles
-
Atg23 is a vesicle-tethering protein.Autophagy. 2022 Oct;18(10):2510-2511. doi: 10.1080/15548627.2022.2105107. Epub 2022 Aug 1. Autophagy. 2022. PMID: 35867625 Free PMC article.
-
Atg23 is essential for the cytoplasm to vacuole targeting pathway and efficient autophagy but not pexophagy.J Biol Chem. 2003 Nov 28;278(48):48445-52. doi: 10.1074/jbc.M309238200. Epub 2003 Sep 22. J Biol Chem. 2003. PMID: 14504273 Free PMC article.
-
Atg23 and Atg27 act at the early stages of Atg9 trafficking in S. cerevisiae.Traffic. 2015 Feb;16(2):172-90. doi: 10.1111/tra.12240. Epub 2014 Dec 16. Traffic. 2015. PMID: 25385507 Free PMC article.
-
Mechanistic Insights into the Role of Atg11 in Selective Autophagy.J Mol Biol. 2020 Jan 3;432(1):104-122. doi: 10.1016/j.jmb.2019.06.017. Epub 2019 Jun 22. J Mol Biol. 2020. PMID: 31238043 Free PMC article. Review.
-
Atg9 trafficking in the yeast Saccharomyces cerevisiae.Autophagy. 2007 Mar-Apr;3(2):145-8. doi: 10.4161/auto.3608. Epub 2007 Mar 21. Autophagy. 2007. PMID: 17204846 Review.
Cited by
-
Characterization of Protein-Membrane Interactions in Yeast Autophagy.Cells. 2022 Jun 9;11(12):1876. doi: 10.3390/cells11121876. Cells. 2022. PMID: 35741004 Free PMC article. Review.
-
Osmotic Pressure Enables High-Yield Assembly of Giant Vesicles in Solutions of Physiological Ionic Strengths.Langmuir. 2023 Apr 18;39(15):5579-5590. doi: 10.1021/acs.langmuir.3c00457. Epub 2023 Apr 6. Langmuir. 2023. PMID: 37021722 Free PMC article.
-
Reliable Approach for Pure Yeast Cell Wall Protein Isolation from Saccharomyces cerevisiae Yeast Cells.ACS Omega. 2022 Aug 15;7(34):29702-29713. doi: 10.1021/acsomega.2c02176. eCollection 2022 Aug 30. ACS Omega. 2022. PMID: 36061670 Free PMC article.
-
Huntingtin contains an ubiquitin-binding domain and regulates lysosomal targeting of mitochondrial and RNA-binding proteins.Proc Natl Acad Sci U S A. 2024 Aug 6;121(32):e2319091121. doi: 10.1073/pnas.2319091121. Epub 2024 Jul 29. Proc Natl Acad Sci U S A. 2024. PMID: 39074279 Free PMC article.
-
Theater in the Self-Cleaning Cell: Intrinsically Disordered Proteins or Protein Regions Acting with Membranes in Autophagy.Membranes (Basel). 2022 Apr 24;12(5):457. doi: 10.3390/membranes12050457. Membranes (Basel). 2022. PMID: 35629783 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

