Identification of a combined apoptosis and hypoxia gene signature for predicting prognosis and immune infiltration in breast cancer
- PMID: 35441810
- PMCID: PMC9582692
- DOI: 10.1002/cam4.4755
Identification of a combined apoptosis and hypoxia gene signature for predicting prognosis and immune infiltration in breast cancer
Abstract
Background: Breast cancer (BC) is the most common malignant tumor worldwide. Apoptosis and hypoxia are involved in the progression of BC, but reliable biomarkers for these have not been developed. We hope to explore a gene signature that combined apoptosis and hypoxia-related genes (AHGs) to predict BC prognosis and immune infiltration.
Methods: We collected the mRNA expression profiles and clinical data information of BC patients from The Cancer Genome Atlas database. The gene signature based on AHGs was constructed using the univariate Cox regression, least absolute shrinkage and selection operator, and multivariate Cox regression analysis. The associations between risk scores, immune infiltration, and immune checkpoint gene expression were studied using single-sample gene set enrichment analysis. Besides, gene signature and independent clinicopathological characteristics were combined to establish a nomogram. Finally, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) were performed on the potential functions of AHGs.
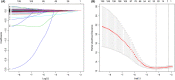
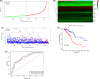
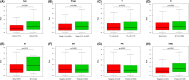
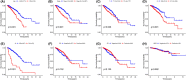
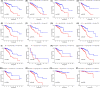
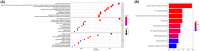
Results: We identified a 16-AHG signature (AGPAT1, BTBD6, EIF4EBP1, ERRFI1, FAM114A1, GRIP1, IRF2, JAK1, MAP2K6, MCTS1, NFKBIA, NFKBIZ, NUP43, PGK1, RCL1, and SGCE) that could independently predict BC prognosis. The median score of the risk model divided the patients into two subgroups. By contrast, patients in the high-risk group had poorer prognosis, less abundance of immune cell infiltration, and expression of immune checkpoint genes. The gene signature and nomogram had good predictive effects on the overall survival of BC patients. GO and KEGG analyses revealed that the differential expression of AHGs may be closely related to tumor immunity.
Conclusion: We established and verified a 16-AHG BC signature which may help predict prognosis, assess potential immunotherapy benefits, and provide inspiration for future research on the functions and mechanisms of AHGs in BC.
Keywords: breast cancer; cancer genetics; immunology; microenvironment; prognosis; risk model.
© 2022 The Authors. Cancer Medicine published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




Similar articles
-
Establishment and validation of an aging-related risk signature associated with prognosis and tumor immune microenvironment in breast cancer.Eur J Med Res. 2022 Dec 29;27(1):317. doi: 10.1186/s40001-022-00924-4. Eur J Med Res. 2022. PMID: 36581948 Free PMC article.
-
A combined hypoxia and immune gene signature for predicting survival and risk stratification in triple-negative breast cancer.Aging (Albany NY). 2021 Aug 2;13(15):19486-19509. doi: 10.18632/aging.203360. Epub 2021 Aug 2. Aging (Albany NY). 2021. PMID: 34341184 Free PMC article.
-
Oncogenic signaling pathway-related long non-coding RNAs for predicting prognosis and immunotherapy response in breast cancer.Front Immunol. 2022 Aug 4;13:891175. doi: 10.3389/fimmu.2022.891175. eCollection 2022. Front Immunol. 2022. PMID: 35990668 Free PMC article.
-
Prediction of Overall Survival Among Female Patients With Breast Cancer Using a Prognostic Signature Based on 8 DNA Repair-Related Genes.JAMA Netw Open. 2020 Oct 1;3(10):e2014622. doi: 10.1001/jamanetworkopen.2020.14622. JAMA Netw Open. 2020. PMID: 33017027 Free PMC article.
-
A novel lactate metabolism-related signature predicts prognosis and tumor immune microenvironment of breast cancer.Front Genet. 2022 Sep 7;13:934830. doi: 10.3389/fgene.2022.934830. eCollection 2022. Front Genet. 2022. PMID: 36171887 Free PMC article.
Cited by
-
Advances in the understanding of nuclear pore complexes in human diseases.J Cancer Res Clin Oncol. 2024 Jul 30;150(7):374. doi: 10.1007/s00432-024-05881-5. J Cancer Res Clin Oncol. 2024. PMID: 39080077 Free PMC article. Review.
-
Construction of an oxidative stress-associated genes signature in breast cancer by machine learning algorithms.J Int Med Res. 2024 Mar;52(3):3000605241232560. doi: 10.1177/03000605241232560. J Int Med Res. 2024. PMID: 38520254 Free PMC article.
-
Development and validation of a prognostic nomogram for breast cancer patients who underwent chemoradiotherapy and surgery: a retrospective cohort study based on the SEER database and two Chinese cohorts.Am J Cancer Res. 2023 Nov 15;13(11):5065-5081. eCollection 2023. Am J Cancer Res. 2023. PMID: 38058820 Free PMC article.
-
NUP43 promotes PD-L1/nPD-L1/PD-L1 feedback loop via TM4SF1/JAK/STAT3 pathway in colorectal cancer progression and metastatsis.Cell Death Discov. 2024 May 18;10(1):241. doi: 10.1038/s41420-024-02025-z. Cell Death Discov. 2024. PMID: 38762481 Free PMC article.
-
Construction of a Novel Disulfidptosis-Related lncRNA Prognostic Signature in Pancreatic Cancer.Mol Biotechnol. 2024 Sep;66(9):2396-2414. doi: 10.1007/s12033-023-00875-z. Epub 2023 Sep 21. Mol Biotechnol. 2024. PMID: 37733182
References
-
- Sung H, Ferlay J, Siegel RL. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. 2021;71(3):209‐249. - PubMed
-
- Siegel RL, Miller KD. Cancer statistics, 2022. 2021;72(1):7‐33. - PubMed
-
- D'Arcy MS. Cell death: a review of the major forms of apoptosis, necrosis and autophagy. Cell Biol Int. 2019;43(6):582‐592. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

