Engineered plant control of associative nitrogen fixation
- PMID: 35412890
- PMCID: PMC9169844
- DOI: 10.1073/pnas.2117465119
Engineered plant control of associative nitrogen fixation
Abstract
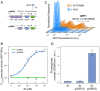
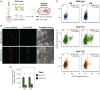
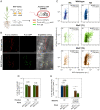
Engineering N2-fixing symbioses between cereals and diazotrophic bacteria represents a promising strategy to sustainably deliver biologically fixed nitrogen (N) in agriculture. We previously developed novel transkingdom signaling between plants and bacteria, through plant production of the bacterial signal rhizopine, allowing control of bacterial gene expression in association with the plant. Here, we have developed both a homozygous rhizopine producing (RhiP) barley line and a hybrid rhizopine uptake system that conveys upon our model bacterium Azorhizobium caulinodans ORS571 (Ac) 103-fold improved sensitivity for rhizopine perception. Using this improved genetic circuitry, we established tight rhizopine-dependent transcriptional control of the nitrogenase master regulator nifA and the N metabolism σ-factor rpoN, which drove nitrogenase expression and activity in vitro and in situ by bacteria colonizing RhiP barley roots. Although in situ nitrogenase activity was suboptimally effective relative to the wild-type strain, activation was specific to RhiP barley and was not observed on the roots of wild-type plants. This work represents a key milestone toward the development of a synthetic plant-controlled symbiosis in which the bacteria fix N2 only when in contact with the desired host plant and are prevented from interaction with nontarget plant species.
Keywords: barley; nitrogen fixation; rhizobium; rhizopine; symbiosis.
Conflict of interest statement
The authors declare no competing interest.
Figures

Similar articles
-
Control of nitrogen fixation and ammonia excretion in Azorhizobium caulinodans.PLoS Genet. 2022 Jun 21;18(6):e1010276. doi: 10.1371/journal.pgen.1010276. eCollection 2022 Jun. PLoS Genet. 2022. PMID: 35727841 Free PMC article.
-
Rhizopine biosensors for plant-dependent control of bacterial gene expression.Environ Microbiol. 2023 Feb;25(2):383-396. doi: 10.1111/1462-2920.16288. Epub 2022 Dec 4. Environ Microbiol. 2023. PMID: 36428208 Free PMC article.
-
Control of nitrogen fixation in bacteria that associate with cereals.Nat Microbiol. 2020 Feb;5(2):314-330. doi: 10.1038/s41564-019-0631-2. Epub 2019 Dec 16. Nat Microbiol. 2020. PMID: 31844298 Free PMC article.
-
Azospirillum, a free-living nitrogen-fixing bacterium closely associated with grasses: genetic, biochemical and ecological aspects.FEMS Microbiol Rev. 2000 Oct;24(4):487-506. doi: 10.1111/j.1574-6976.2000.tb00552.x. FEMS Microbiol Rev. 2000. PMID: 10978548 Review.
-
Nitrogen signalling in plant interactions with associative and endophytic diazotrophic bacteria.J Exp Bot. 2014 Oct;65(19):5631-42. doi: 10.1093/jxb/eru319. Epub 2014 Aug 11. J Exp Bot. 2014. PMID: 25114015 Review.
Cited by
-
Climate change challenges, plant science solutions.Plant Cell. 2023 Jan 2;35(1):24-66. doi: 10.1093/plcell/koac303. Plant Cell. 2023. PMID: 36222573 Free PMC article.
-
Comparative phylotranscriptomics reveals ancestral and derived root nodule symbiosis programmes.Nat Plants. 2023 Jul;9(7):1067-1080. doi: 10.1038/s41477-023-01441-w. Epub 2023 Jun 15. Nat Plants. 2023. PMID: 37322127 Free PMC article.
-
Biological nitrogen fixation in cereal crops: Progress, strategies, and perspectives.Plant Commun. 2023 Mar 13;4(2):100499. doi: 10.1016/j.xplc.2022.100499. Epub 2022 Nov 28. Plant Commun. 2023. PMID: 36447432 Free PMC article. Review.
-
Diurnal switches in diazotrophic lifestyle increase nitrogen contribution to cereals.Nat Commun. 2023 Nov 18;14(1):7516. doi: 10.1038/s41467-023-43370-4. Nat Commun. 2023. PMID: 37980355 Free PMC article.
-
Molecular Mechanism and Agricultural Application of the NifA-NifL System for Nitrogen Fixation.Int J Mol Sci. 2023 Jan 4;24(2):907. doi: 10.3390/ijms24020907. Int J Mol Sci. 2023. PMID: 36674420 Free PMC article. Review.
References
-
- O’Hara G. W., The role of nitrogen fixation in crop production. J. Crop Prod. 1, 115–138 (1998).
-
- Herridge D. F., Peoples M. B., Boddey R. M., Global inputs of biological nitrogen fixation in agricultural systems. Plant Soil 311, 1–18 (2008).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

