Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19
- PMID: 35393402
- PMCID: PMC8989888
- DOI: 10.1038/s41467-022-29413-2
Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19
Abstract
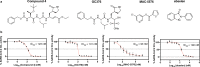
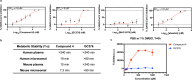
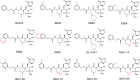
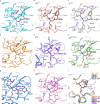
The SARS-CoV-2 3CL protease is a critical drug target for small molecule COVID-19 therapy, given its likely druggability and essentiality in the viral maturation and replication cycle. Based on the conservation of 3CL protease substrate binding pockets across coronaviruses and using screening, we identified four structurally distinct lead compounds that inhibit SARS-CoV-2 3CL protease. After evaluation of their binding specificity, cellular antiviral potency, metabolic stability, and water solubility, we prioritized the GC376 scaffold as being optimal for optimization. We identified multiple drug-like compounds with <10 nM potency for inhibiting SARS-CoV-2 3CL and the ability to block SARS-CoV-2 replication in human cells, obtained co-crystal structures of the 3CL protease in complex with these compounds, and determined that they have pan-coronavirus activity. We selected one compound, termed coronastat, as an optimized lead and characterized it in pharmacokinetic and safety studies in vivo. Coronastat represents a new candidate for a small molecule protease inhibitor for the treatment of SARS-CoV-2 infection for eliminating pandemics involving coronaviruses.
© 2022. The Author(s).
Conflict of interest statement
S.I., H.L., A.Z., B.R.S., A.C., T.R., N.K., E.B., F.F., N.E.S.T., S.L., and D.D.H. are inventors on invention disclosures and patent applications submitted based on this work. B.R.S. is an inventor on additional patents and patent applications related to small molecule therapeutics, and co-founded and serves as a consultant to Inzen Therapeutics, Nevrox Limited, Exarta Therapeutics, and ProJenX Inc., and serves as a consultant to Weatherwax Biotechnologies and Akin Gump Strauss Hauer & Feld LLP. A.Z. is an inventor on additional patents and patent applications related to small molecule therapeutics, and co-founded and serves as a consultant to ProJenX Inc. C.Q., W.L., H.S., C.J., M.A.L., and T.M. are employed by Waters Corporation. The remaining authors declare no competing interests.
Figures

Similar articles
-
Design and Evaluation of Anti-SARS-Coronavirus Agents Based on Molecular Interactions with the Viral Protease.Molecules. 2020 Aug 27;25(17):3920. doi: 10.3390/molecules25173920. Molecules. 2020. PMID: 32867349 Free PMC article. Review.
-
N-Terminal Finger Stabilizes the S1 Pocket for the Reversible Feline Drug GC376 in the SARS-CoV-2 Mpro Dimer.J Mol Biol. 2021 Jun 25;433(13):167003. doi: 10.1016/j.jmb.2021.167003. Epub 2021 Apr 22. J Mol Biol. 2021. PMID: 33895266 Free PMC article.
-
In silico Study to Evaluate the Antiviral Activity of Novel Structures against 3C-like Protease of Novel Coronavirus (COVID-19) and SARS-CoV.Med Chem. 2021;17(4):380-395. doi: 10.2174/1573396316999200727125522. Med Chem. 2021. PMID: 32720605
-
Potential of coronavirus 3C-like protease inhibitors for the development of new anti-SARS-CoV-2 drugs: Insights from structures of protease and inhibitors.Int J Antimicrob Agents. 2020 Aug;56(2):106055. doi: 10.1016/j.ijantimicag.2020.106055. Epub 2020 Jun 11. Int J Antimicrob Agents. 2020. PMID: 32534187 Free PMC article.
-
Overview of antiviral drug candidates targeting coronaviral 3C-like main proteases.FEBS J. 2021 Sep;288(17):5089-5121. doi: 10.1111/febs.15696. Epub 2021 Feb 1. FEBS J. 2021. PMID: 33400393 Review.
Cited by
-
SARS-CoV-2 SUD2 and Nsp5 Conspire to Boost Apoptosis of Respiratory Epithelial Cells via an Augmented Interaction with the G-Quadruplex of BclII.mBio. 2023 Apr 25;14(2):e0335922. doi: 10.1128/mbio.03359-22. Epub 2023 Feb 28. mBio. 2023. PMID: 36853058 Free PMC article.
-
Understanding Cysteine Chemistry Using Conventional and Serial X-Ray Protein Crystallography.Crystals (Basel). 2022 Nov;12(11):1671. doi: 10.3390/cryst12111671. Epub 2022 Nov 19. Crystals (Basel). 2022. PMID: 36685087 Free PMC article.
-
Potential Anti-SARS-CoV-2 Prodrugs Activated by Phosphorylation and Their Role in the Aged Population.Molecules. 2023 Mar 2;28(5):2332. doi: 10.3390/molecules28052332. Molecules. 2023. PMID: 36903575 Free PMC article. Review.
-
Design, synthesis, and biochemical and computational screening of novel oxindole derivatives as inhibitors of Aurora A kinase and SARS-CoV-2 spike/host ACE2 interaction.Med Chem Res. 2024;33(4):620-634. doi: 10.1007/s00044-024-03201-7. Epub 2024 Mar 5. Med Chem Res. 2024. PMID: 38646411 Free PMC article.
-
Functional map of SARS-CoV-2 3CL protease reveals tolerant and immutable sites.Cell Host Microbe. 2022 Oct 12;30(10):1354-1362.e6. doi: 10.1016/j.chom.2022.08.003. Epub 2022 Aug 11. Cell Host Microbe. 2022. PMID: 36029764 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

