DNA circles promote yeast ageing in part through stimulating the reorganization of nuclear pore complexes
- PMID: 35373738
- PMCID: PMC9020822
- DOI: 10.7554/eLife.71196
DNA circles promote yeast ageing in part through stimulating the reorganization of nuclear pore complexes
Abstract
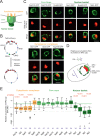
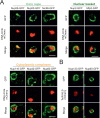
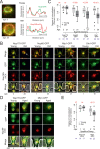
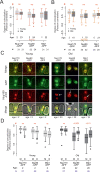
The nuclear pore complex (NPC) mediates nearly all exchanges between nucleus and cytoplasm, and in many species, it changes composition as the organism ages. However, how these changes arise and whether they contribute themselves to ageing is poorly understood. We show that SAGA-dependent attachment of DNA circles to NPCs in replicatively ageing yeast cells causes NPCs to lose their nuclear basket and cytoplasmic complexes. These NPCs were not recognized as defective by the NPC quality control machinery (SINC) and not targeted by ESCRTs. They interacted normally or more effectively with protein import and export factors but specifically lost mRNA export factors. Acetylation of Nup60 drove the displacement of basket and cytoplasmic complexes from circle-bound NPCs. Mutations preventing this remodeling extended the replicative lifespan of the cells. Thus, our data suggest that the anchorage of accumulating circles locks NPCs in a specialized state and that this process is intrinsically linked to the mechanisms by which ERCs promote ageing.
Keywords: DNA circles; NPC; S. cerevisiae; aging; cell biology; evolutionary biology; nuclear basket.
© 2022, Meinema et al.
Conflict of interest statement
AM, AM, MM, TA, SL, GC, YB No competing interests declared
Figures

Similar articles
-
Nuclear pore basket proteins are tethered to the nuclear envelope and can regulate membrane curvature.Dev Cell. 2015 May 4;33(3):285-98. doi: 10.1016/j.devcel.2015.02.017. Dev Cell. 2015. PMID: 25942622 Free PMC article.
-
Nuclear pore complex acetylation regulates mRNA export and cell cycle commitment in budding yeast.EMBO J. 2022 Aug 1;41(15):e110271. doi: 10.15252/embj.2021110271. Epub 2022 Jun 23. EMBO J. 2022. PMID: 35735140 Free PMC article.
-
Role of SAGA in the asymmetric segregation of DNA circles during yeast ageing.Elife. 2014 Nov 17;3:e03790. doi: 10.7554/eLife.03790. Elife. 2014. PMID: 25402830 Free PMC article.
-
[Nuclear pores: from yeast to higher eukaryotes].J Soc Biol. 2002;196(4):349-54. J Soc Biol. 2002. PMID: 12645306 Review. French.
-
Nuclear pore complex composition: a new regulator of tissue-specific and developmental functions.Nat Rev Mol Cell Biol. 2012 Nov;13(11):687-99. doi: 10.1038/nrm3461. Nat Rev Mol Cell Biol. 2012. PMID: 23090414 Review.
Cited by
-
Dietary change without caloric restriction maintains a youthful profile in ageing yeast.PLoS Biol. 2023 Aug 29;21(8):e3002245. doi: 10.1371/journal.pbio.3002245. eCollection 2023 Aug. PLoS Biol. 2023. PMID: 37643155 Free PMC article.
-
Senescence in yeast is associated with amplified linear fragments of chromosome XII rather than ribosomal DNA circle accumulation.PLoS Biol. 2023 Aug 29;21(8):e3002250. doi: 10.1371/journal.pbio.3002250. eCollection 2023 Aug. PLoS Biol. 2023. PMID: 37643194 Free PMC article.
-
Quantitative analysis of nuclear pore complex organization in Schizosaccharomyces pombe.Life Sci Alliance. 2022 Mar 30;5(7):e202201423. doi: 10.26508/lsa.202201423. Print 2022 Jul. Life Sci Alliance. 2022. PMID: 35354597 Free PMC article.
-
The Nuclear Pore Complex: Birth, Life, and Death of a Cellular Behemoth.Cells. 2022 Apr 25;11(9):1456. doi: 10.3390/cells11091456. Cells. 2022. PMID: 35563762 Free PMC article. Review.
-
High-fidelity (repeat) consensus sequences from short reads using combined read clustering and assembly.BMC Genomics. 2024 Jan 24;25(1):109. doi: 10.1186/s12864-023-09948-4. BMC Genomics. 2024. PMID: 38267856 Free PMC article.
References
-
- Akey CW, Singh D, Ouch C, Echeverria I, Nudelman I, Varberg JM, Yu Z, Fang F, Shi Y, Wang J, Salzberg D, Song K, Xu C, Gumbart JC, Suslov S, Unruh J, Jaspersen SL, Chait BT, Sali A, Fernandez-Martinez J, Ludtke SJ, Villa E, Rout MP. Comprehensive structure and functional adaptations of the yeast nuclear pore complex. Cell. 2022;185:361–378. doi: 10.1016/j.cell.2021.12.015. - DOI - PMC - PubMed
-
- Allegretti M, Zimmerli CE, Rantos V, Wilfling F, Ronchi P, Fung HKH, Lee CW, Hagen W, Turoňová B, Karius K, Börmel M, Zhang X, Müller CW, Schwab Y, Mahamid J, Pfander B, Kosinski J, Beck M. In-cell architecture of the nuclear pore and snapshots of its turnover. Nature. 2020;586:796–800. doi: 10.1038/s41586-020-2670-5. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

