Novel Anti-LY6G6D/CD3 T-Cell-Dependent Bispecific Antibody for the Treatment of Colorectal Cancer
- PMID: 35364611
- PMCID: PMC9381132
- DOI: 10.1158/1535-7163.MCT-21-0599
Novel Anti-LY6G6D/CD3 T-Cell-Dependent Bispecific Antibody for the Treatment of Colorectal Cancer
Abstract
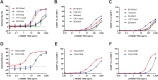
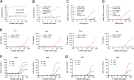
New therapeutics and combination regimens have led to marked clinical improvements for the treatment of a subset of colorectal cancer. Immune checkpoint inhibitors have shown clinical efficacy in patients with mismatch-repair-deficient or microsatellite instability-high (MSI-H) metastatic colorectal cancer (mCRC). However, patients with microsatellite-stable (MSS) or low levels of microsatellite instable (MSI-L) colorectal cancer have not benefited from these immune modulators, and the survival outcome remains poor for the majority of patients diagnosed with mCRC. In this article, we describe the discovery of a novel T-cell-dependent bispecific antibody (TDB) targeting tumor-associated antigen LY6G6D, LY6G6D-TDB, for the treatment of colorectal cancer. RNAseq analysis showed that LY6G6D was differentially expressed in colorectal cancer with high prevalence in MSS and MSI-L subsets, whereas LY6G6D expression in normal tissues was limited. IHC confirmed the elevated expression of LY6G6D in primary and metastatic colorectal tumors, whereas minimal or no expression was observed in most normal tissue samples. The optimized LY6G6D-TDB, which targets a membrane-proximal epitope of LY6G6D and binds to CD3 with high affinity, exhibits potent antitumor activity both in vitro and in vivo. In vitro functional assays show that LY6G6D-TDB-mediated T-cell activation and cytotoxicity are conditional and target dependent. In mouse xenograft tumor models, LY6G6D-TDB demonstrates antitumor efficacy as a single agent against established colorectal tumors, and enhanced efficacy can be achieved when LY6G6D-TDB is combined with PD-1 blockade. Our studies provide evidence for the therapeutic potential of LY6G6D-TDB as an effective treatment option for patients with colorectal cancer.
©2022 The Authors; Published by the American Association for Cancer Research.
Figures




Similar articles
-
LY6G6D is a selectively expressed colorectal cancer antigen that can be used for targeting a therapeutic T-cell response by a T-cell engager.Front Immunol. 2022 Sep 8;13:1008764. doi: 10.3389/fimmu.2022.1008764. eCollection 2022. Front Immunol. 2022. PMID: 36159851 Free PMC article.
-
JAK/Stat5-mediated subtype-specific lymphocyte antigen 6 complex, locus G6D (LY6G6D) expression drives mismatch repair proficient colorectal cancer.J Exp Clin Cancer Res. 2019 Jan 22;38(1):28. doi: 10.1186/s13046-018-1019-5. J Exp Clin Cancer Res. 2019. PMID: 30670049 Free PMC article.
-
IFNγ-induced Chemokines Are Required for CXCR3-mediated T-Cell Recruitment and Antitumor Efficacy of Anti-HER2/CD3 Bispecific Antibody.Clin Cancer Res. 2018 Dec 15;24(24):6447-6458. doi: 10.1158/1078-0432.CCR-18-1139. Epub 2018 Jun 27. Clin Cancer Res. 2018. PMID: 29950350
-
Latest evidence on immune checkpoint inhibitors in metastatic colorectal cancer: A 2022 update.Crit Rev Oncol Hematol. 2022 May;173:103663. doi: 10.1016/j.critrevonc.2022.103663. Epub 2022 Mar 26. Crit Rev Oncol Hematol. 2022. PMID: 35351582 Review.
-
Immune Checkpoint Inhibition in Colorectal Cancer: Microsatellite Instability and Beyond.Target Oncol. 2020 Feb;15(1):11-24. doi: 10.1007/s11523-019-00690-0. Target Oncol. 2020. PMID: 31786718 Review.
Cited by
-
LY6G6D is a selectively expressed colorectal cancer antigen that can be used for targeting a therapeutic T-cell response by a T-cell engager.Front Immunol. 2022 Sep 8;13:1008764. doi: 10.3389/fimmu.2022.1008764. eCollection 2022. Front Immunol. 2022. PMID: 36159851 Free PMC article.
-
Clinical and Immunologic Characteristics of Colorectal Cancer Tumors Expressing LY6G6D.Int J Mol Sci. 2024 May 14;25(10):5345. doi: 10.3390/ijms25105345. Int J Mol Sci. 2024. PMID: 38791382 Free PMC article.
-
Spatially resolved transcriptomics revealed local invasion-related genes in colorectal cancer.Front Oncol. 2023 Feb 1;13:1089090. doi: 10.3389/fonc.2023.1089090. eCollection 2023. Front Oncol. 2023. PMID: 36816947 Free PMC article.
-
Considerations for the clinical development of immuno-oncology agents in cancer.Front Immunol. 2023 Aug 11;14:1229575. doi: 10.3389/fimmu.2023.1229575. eCollection 2023. Front Immunol. 2023. PMID: 37638048 Free PMC article. Review.
References
-
- GLOBOCAN database, 2020: Available from:http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx#.
-
- Siegel RL, Miller KD, Sauer AG, Fedewa SA, Butterly LF, Anderson JC, et al. . Colorectal cancer statistics, 2020. CA Cancer J Clin 2020;70:145–64. - PubMed
-
- Overman MJ, Mcdermott R, Leach JL, Lonardi S, Lenz H-J, Morse MA, et al. . Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol 2017;18:1182–91. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

