IL-1β-activated mTORC2 promotes accumulation of IFN-γ+ γδ T cells by upregulating CXCR3 to restrict hepatic fibrosis
- PMID: 35361750
- PMCID: PMC8971410
- DOI: 10.1038/s41419-022-04739-3
IL-1β-activated mTORC2 promotes accumulation of IFN-γ+ γδ T cells by upregulating CXCR3 to restrict hepatic fibrosis
Abstract
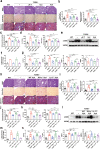
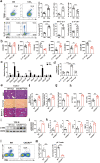
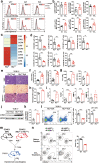
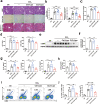
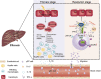
Liver fibrosis represents a severe stage of liver damage, with hallmarks of inflammation, hepatic stellate cell activation, and extracellular matrix accumulation. Although previous studies demonstrated γδ T cells are involved in liver fibrosis, the precise role and mechanisms of γδ T cells migrating to fibrotic liver have not been elucidated. Here, we aim to investigate the functional subsets of γδ T cells in hepatic fibrosis and to further explore the underlying causes and drivers of migration. In this study, we observed that γδ T cells accumulate in fibrotic liver. Adoptive transfer of γδ T, especially Vγ4 γδ T subset, can significantly alleviate liver fibrosis. In addition, CCl4 treatment also leads to activation of mTOR signaling in γδ T cells. Genetic deletion of the Rictor gene, but not Raptor, in γδ T cells markedly exacerbated liver fibrosis. Mechanistically, CCl4-induced liver injury causes macrophage accumulation in the liver, and IL-1β produced by macrophages promotes mTORC2 signaling activation in γδ T cells, which upregulates T-bet expression and eventually promotes CXCR3 transcription to drive γδ T cell migration. Moreover, hepatic γδ T cells ameliorated liver fibrosis by cytotoxicity against activated hepatic stellate cells in FasL-dependent manner, and secrete IFN-γ to inhibit the differentiation of pro-fibrotic Th17 cells. Thus, IL-1β-activated mTORC2 signaling in γδ T cells upregulates CXCR3 expression, which is critical for IFN-γ+ γδ T cells migration into the liver and amelioration of liver fibrosis. Our findings indicate that targeting the mTORC2 or CXCR3 in γδ T cells could be considered as a promising approach for γδ T cell immunotherapy against liver fibrosis.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Chemokine receptor CCR6-dependent accumulation of γδ T cells in injured liver restricts hepatic inflammation and fibrosis.Hepatology. 2014 Feb;59(2):630-42. doi: 10.1002/hep.26697. Epub 2013 Dec 23. Hepatology. 2014. PMID: 23959575 Free PMC article.
-
Exosome-mediated activation of toll-like receptor 3 in stellate cells stimulates interleukin-17 production by γδ T cells in liver fibrosis.Hepatology. 2016 Aug;64(2):616-31. doi: 10.1002/hep.28644. Epub 2016 Jun 29. Hepatology. 2016. PMID: 27178735
-
Roles of mTORC1 and mTORC2 in controlling γδ T1 and γδ T17 differentiation and function.Cell Death Differ. 2020 Jul;27(7):2248-2262. doi: 10.1038/s41418-020-0500-9. Epub 2020 Jan 30. Cell Death Differ. 2020. PMID: 32001780 Free PMC article.
-
Cross-talk between hepatic stellate cells and T lymphocytes in liver fibrosis.Hepatobiliary Pancreat Dis Int. 2021 Jun;20(3):207-214. doi: 10.1016/j.hbpd.2021.04.007. Epub 2021 Apr 28. Hepatobiliary Pancreat Dis Int. 2021. PMID: 33972160 Review.
-
The Role of γδ T Cells in Fibrotic Diseases.Rambam Maimonides Med J. 2016 Oct 31;7(4):e0029. doi: 10.5041/RMMJ.10256. Rambam Maimonides Med J. 2016. PMID: 27824548 Free PMC article. Review.
Cited by
-
Macrophage plasticity: signaling pathways, tissue repair, and regeneration.MedComm (2020). 2024 Aug 1;5(8):e658. doi: 10.1002/mco2.658. eCollection 2024 Aug. MedComm (2020). 2024. PMID: 39092292 Free PMC article. Review.
-
Ongoing involvers and promising therapeutic targets of hepatic fibrosis: The hepatic immune microenvironment.Front Immunol. 2023 Feb 16;14:1131588. doi: 10.3389/fimmu.2023.1131588. eCollection 2023. Front Immunol. 2023. PMID: 36875101 Free PMC article. Review.
-
γδ T cell-mediated activation of cDC1 orchestrates CD4+ Th1 cell priming in malaria.Front Immunol. 2024 Aug 15;15:1426316. doi: 10.3389/fimmu.2024.1426316. eCollection 2024. Front Immunol. 2024. PMID: 39211036 Free PMC article.
-
Orchestrated regulation of immune inflammation with cell therapy in pediatric acute liver injury.Front Immunol. 2023 Jun 22;14:1194588. doi: 10.3389/fimmu.2023.1194588. eCollection 2023. Front Immunol. 2023. PMID: 37426664 Free PMC article. Review.
-
Rictor/mTORC2 signalling contributes to renal vascular endothelial-to-mesenchymal transition and renal allograft interstitial fibrosis by regulating BNIP3-mediated mitophagy.Clin Transl Med. 2024 May;14(5):e1686. doi: 10.1002/ctm2.1686. Clin Transl Med. 2024. PMID: 38769658 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

