Selective covalent capture of collagen triple helices with a minimal protecting group strategy
- PMID: 35356674
- PMCID: PMC8890135
- DOI: 10.1039/d1sc06361h
Selective covalent capture of collagen triple helices with a minimal protecting group strategy
Abstract
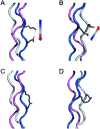
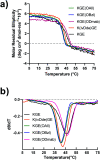
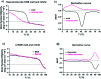
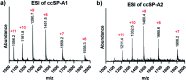
Collagens and their most characteristic structural unit, the triple helix, play many critical roles in living systems which drive interest in preparing mimics of them. However, application of collagen mimetic helices is limited by poor thermal stability, slow rates of folding and poor equilibrium between monomer and trimer. Covalent capture of the self-assembled triple helix can solve these problems while preserving the native three-dimensional structure critical for biological function. Covalent capture takes advantage of strategically placed lysine and glutamate (or aspartate) residues which form stabilizing charge-pair interactions in the supramolecular helix and can subsequently be converted to isopeptide amide bonds under folded, aqueous conditions. While covalent capture is powerful, charge paired residues are frequently found in natural sequences which must be preserved to maintain biological function. Here we describe a minimal protecting group strategy to allow selective covalent capture of specific charge paired residues which leaves other charged residues unaltered. We investigate a series of side chain protecting groups for lysine and glutamate in model peptides for their ability to be deprotected easily and in high yield while maintaining (1) the solubility of the peptides in water, (2) the self-assembly and stability of the triple helix, and (3) the ability to covalently capture unprotected charge pairs. Optimized conditions are then illustrated in peptides derived from Pulmonary Surfactant protein A (SP-A). These covalently captured SP-A triple helices are found to have dramatically improved rates of folding and thermal stability while maintaining unmodified lysine-glutamate pairs in addition to other unmodified chemical functionality. The approach we illustrate allows for the covalent capture of collagen-like triple helices with virtually any sequence, composition or register. This dramatically broadens the utility of the covalent capture approach to the stabilization of biomimetic triple helices and thus also improves the utility of biomimetic collagens generally.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures



Similar articles
-
Covalent Capture of Collagen Triple Helices Using Lysine-Aspartate and Lysine-Glutamate Pairs.Biomacromolecules. 2020 Sep 14;21(9):3772-3781. doi: 10.1021/acs.biomac.0c00878. Epub 2020 Aug 21. Biomacromolecules. 2020. PMID: 32820897
-
Stabilization of Synthetic Collagen Triple Helices: Charge Pairs and Covalent Capture.Biomacromolecules. 2023 Nov 13;24(11):5083-5090. doi: 10.1021/acs.biomac.3c00680. Epub 2023 Oct 23. Biomacromolecules. 2023. PMID: 37871141
-
Covalent Capture of a Collagen Mimetic Peptide with an Integrin-Binding Motif.Biomacromolecules. 2022 Jun 13;23(6):2396-2403. doi: 10.1021/acs.biomac.2c00155. Epub 2022 Apr 21. Biomacromolecules. 2022. PMID: 35446536
-
Pairwise interactions in collagen and the design of heterotrimeric helices.Curr Opin Chem Biol. 2013 Dec;17(6):960-7. doi: 10.1016/j.cbpa.2013.10.019. Epub 2013 Nov 16. Curr Opin Chem Biol. 2013. PMID: 24252327 Review.
-
Triple-helical peptides: an approach to collagen conformation, stability, and self-association.Biopolymers. 2008 May;89(5):345-53. doi: 10.1002/bip.20958. Biopolymers. 2008. PMID: 18275087 Review.
Cited by
-
Beyond the Triple Helix: Exploration of the Hierarchical Assembly Space of Collagen-like Peptides.bioRxiv [Preprint]. 2024 May 15:2024.05.14.594194. doi: 10.1101/2024.05.14.594194. bioRxiv. 2024. Update in: Nat Commun. 2024 Nov 29;15(1):10385. doi: 10.1038/s41467-024-54560-z. PMID: 38798367 Free PMC article. Updated. Preprint.
References
-
- Guo Z.-Y. Feng Y.-M. Biol. Chem. 2001;382:443–448. - PubMed
LinkOut - more resources
Full Text Sources

