Possible Cross-Reactivity of Feline and White-Tailed Deer Antibodies against the SARS-CoV-2 Receptor Binding Domain
- PMID: 35352999
- PMCID: PMC9044950
- DOI: 10.1128/jvi.00250-22
Possible Cross-Reactivity of Feline and White-Tailed Deer Antibodies against the SARS-CoV-2 Receptor Binding Domain
Abstract
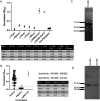
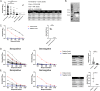
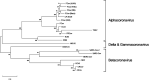
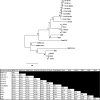
In late 2019, a novel coronavirus began circulating within humans in central China. It was designated SARS-CoV-2 because of its genetic similarities to the 2003 SARS coronavirus (SARS-CoV). Now that SARS-CoV-2 has spread worldwide, there is a risk of it establishing new animal reservoirs and recombination with native circulating coronaviruses. To screen local animal populations in the United States for exposure to SARS-like coronaviruses, we developed a serological assay using the receptor binding domain (RBD) from SARS-CoV-2. SARS-CoV-2's RBD is antigenically distinct from common human and animal coronaviruses, allowing us to identify animals previously infected with SARS-CoV or SARS-CoV-2. Using an indirect enzyme-linked immunosorbent assay (ELISA) for SARS-CoV-2's RBD, we screened serum from wild and domestic animals for the presence of antibodies against SARS-CoV-2's RBD. Surprisingly prepandemic feline serum samples submitted to the University of Tennessee Veterinary Hospital were ∼50% positive for anti-SARS RBD antibodies. Some of these samples were serologically negative for feline coronavirus (FCoV), raising the question of the etiological agent generating anti-SARS-CoV-2 RBD cross-reactivity. We also identified several white-tailed deer from South Carolina with anti-SARS-CoV-2 antibodies. These results are intriguing, as cross-reactive antibodies toward SARS-CoV-2 RBD have not been reported to date. The etiological agent responsible for seropositivity was not readily apparent, but finding seropositive cats prior to the current SARS-CoV-2 pandemic highlights our lack of information about circulating coronaviruses in other species. IMPORTANCE We report cross-reactive antibodies from prepandemic cats and postpandemic South Carolina white-tailed deer that are specific for that SARS-CoV RBD. There are several potential explanations for this cross-reactivity, each with important implications to coronavirus disease surveillance. Perhaps the most intriguing possibility is the existence and transmission of an etiological agent (such as another coronavirus) with similarity to SARS-CoV-2's RBD region. However, we lack conclusive evidence of prepandemic transmission of a SARS-like virus. Our findings provide impetus for the adoption of a One Health Initiative focusing on infectious disease surveillance of multiple animal species to predict the next zoonotic transmission to humans and future pandemics.
Keywords: ELISA; RBD; SARS-CoV-2; antibodies; bovine; canine; coronavirus; cross-reactive; feline; white-tailed deer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Both Feline Coronavirus Serotypes 1 and 2 Infected Domestic Cats Develop Cross-Reactive Antibodies to SARS-CoV-2 Receptor Binding Domain: Its Implication to Pan-CoV Vaccine Development.Viruses. 2023 Mar 31;15(4):914. doi: 10.3390/v15040914. Viruses. 2023. PMID: 37112894 Free PMC article.
-
Seroprevalence of SARS-CoV-2 (COVID-19) exposure in pet cats and dogs in Minnesota, USA.Virulence. 2021 Dec;12(1):1597-1609. doi: 10.1080/21505594.2021.1936433. Virulence. 2021. PMID: 34125647 Free PMC article.
-
Detection of Serum Cross-Reactive Antibodies and Memory Response to SARS-CoV-2 in Prepandemic and Post-COVID-19 Convalescent Samples.J Infect Dis. 2021 Oct 28;224(8):1305-1315. doi: 10.1093/infdis/jiab333. J Infect Dis. 2021. PMID: 34161567 Free PMC article.
-
Mind the feline coronavirus: Comparison with SARS-CoV-2.Gene. 2022 May 30;825:146443. doi: 10.1016/j.gene.2022.146443. Epub 2022 Mar 22. Gene. 2022. PMID: 35337854 Free PMC article. Review.
-
Targeting SARS-CoV2 Spike Protein Receptor Binding Domain by Therapeutic Antibodies.Biomed Pharmacother. 2020 Oct;130:110559. doi: 10.1016/j.biopha.2020.110559. Epub 2020 Aug 1. Biomed Pharmacother. 2020. PMID: 32768882 Free PMC article. Review.
Cited by
-
A comparative study of eight serological methods shows that spike protein-based ELISAs are the most accurate tests for serodiagnosing SARS-CoV-2 infections in cats and dogs.Front Vet Sci. 2023 Jan 26;10:1121935. doi: 10.3389/fvets.2023.1121935. eCollection 2023. Front Vet Sci. 2023. PMID: 36777670 Free PMC article.
-
Detection of SARS-CoV-2 Delta Variant (B.1.617.2) in Domestic Dogs and Zoo Tigers in England and Jersey during 2021.Viruses. 2024 Apr 16;16(4):617. doi: 10.3390/v16040617. Viruses. 2024. PMID: 38675958 Free PMC article.
-
Preventing future zoonosis: SARS-CoV-2 mutations enhance human-animal cross-transmission.Comput Biol Med. 2024 Nov;182:109101. doi: 10.1016/j.compbiomed.2024.109101. Epub 2024 Sep 6. Comput Biol Med. 2024. PMID: 39243518
-
Serological Survey of Retrovirus and Coronavirus Infections, including SARS-CoV-2, in Rural Stray Cats in The Netherlands, 2020-2022.Viruses. 2023 Jul 12;15(7):1531. doi: 10.3390/v15071531. Viruses. 2023. PMID: 37515217 Free PMC article.
-
Both Feline Coronavirus Serotypes 1 and 2 Infected Domestic Cats Develop Cross-Reactive Antibodies to SARS-CoV-2 Receptor Binding Domain: Its Implication to Pan-CoV Vaccine Development.Viruses. 2023 Mar 31;15(4):914. doi: 10.3390/v15040914. Viruses. 2023. PMID: 37112894 Free PMC article.
References
-
- Guan W-J, Ni Z-Y, Hu Y, Liang W-H, Ou C-Q, He J-X, Liu L, Shan H, Lei C-L, Hui DSC, Du B, Li L-J, Zeng G, Yuen K-Y, Chen R-C, Tang C-L, Wang T, Chen P-Y, Xiang J, Li S-Y, Wang J-L, Liang Z-J, Peng Y-X, Wei L, Liu Y, Hu Y-H, Peng P, Wang J-M, Liu J-Y, Chen Z, Li G, Zheng Z-J, Qiu S-Q, Luo J, Ye C-J, Zhu S-Y, Zhong N-S, China Medical Treatment Expert Group for Covid-19. 2020. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 382:1708–1720. 10.1056/NEJMoa2002032. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous

