Deep sequencing of the HIV-1 polymerase gene for characterisation of cytotoxic T-lymphocyte epitopes during early and chronic disease stages
- PMID: 35346259
- PMCID: PMC8959563
- DOI: 10.1186/s12985-022-01772-8
Deep sequencing of the HIV-1 polymerase gene for characterisation of cytotoxic T-lymphocyte epitopes during early and chronic disease stages
Erratum in
-
Correction to: Deep sequencing of the HIV-1 polymerase gene for characterisation of cytotoxic T-lymphocyte epitopes during early and chronic disease stages.Virol J. 2022 May 5;19(1):78. doi: 10.1186/s12985-022-01803-4. Virol J. 2022. PMID: 35513827 Free PMC article. No abstract available.
Abstract
Background: Despite multiple attempts, there is still no effective HIV-1 vaccine available. The HIV-1 polymerase (pol) gene is highly conserved and encodes cytotoxic T-lymphocyte (CTL) epitopes. The aim of the study was to characterise HIV-1 Pol CTL epitopes in mostly sample pairs obtained during early and chronic stages of infection.
Methods: Illumina deep sequencing was performed for all samples while Sanger sequencing was only performed on baseline samples. Codons under immune selection pressure were assessed by computing nonsynonymous to synonymous mutation ratios using MEGA. Minority CTL epitope variants occurring at [Formula: see text] 5% were detected using low-frequency variant tool in CLC Genomics. Los Alamos HIV database was used for mapping mutations to known HIV-1 CTL epitopes.
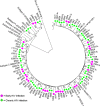
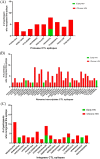
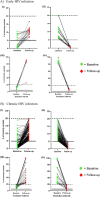
Results: Fifty-two participants were enrolled in the study. Their median age was 28 years (interquartile range: 24-32 years) and majority of participants (92.3%) were female. Illumina minority variant analysis identified a significantly higher number of CTL epitopes (n = 65) compared to epitopes (n = 8) identified through Sanger sequencing. Most of the identified epitopes mapped to reverse transcriptase (RT) and integrase (IN) regardless of sequencing method. There was a significantly higher proportion of minority variant epitopes in RT (n = 39, 60.0%) compared to IN (n = 17, 26.2%) and PR (n = 9, 13.8%), p = 0.002 and < 0.0001, respectively. However, no significant difference was observed between the proportion of minority variant epitopes in IN versus PR, p = 0.06. Some epitopes were detected in either early or chronic HIV-1 infection whereas others were detected in both stages. Different distribution patterns of minority variant epitopes were observed in sample pairs; with some increasing or decreasing over time, while others remained constant. Some of the identified epitopes have not been previously reported for HIV-1 subtype C. There were also variants that could not be mapped to reported CTL epitopes in the Los Alamos HIV database.
Conclusion: Deep sequencing revealed many Pol CTL epitopes, including some not previously reported for HIV-1 subtype C. The findings of this study support the inclusion of RT and IN epitopes in HIV-1 vaccine candidates as these proteins harbour many CTL epitopes.
Keywords: Chronic HIV-1 infection; Early HIV-1 infection; Illumina deep sequencing; Minority variants; Pol CTL epitopes.
© 2022. The Author(s).
Conflict of interest statement
The authors declared no competing interest.
Figures
Similar articles
-
Next-generation sequencing analyses of the emergence and maintenance of mutations in CTL epitopes in HIV controllers with differential viremia control.Retrovirology. 2018 Sep 10;15(1):62. doi: 10.1186/s12977-018-0444-z. Retrovirology. 2018. PMID: 30201008 Free PMC article.
-
Fitness-Balanced Escape Determines Resolution of Dynamic Founder Virus Escape Processes in HIV-1 Infection.J Virol. 2015 Oct;89(20):10303-18. doi: 10.1128/JVI.01876-15. Epub 2015 Jul 29. J Virol. 2015. PMID: 26223634 Free PMC article.
-
Frequent associations between CTL and T-Helper epitopes in HIV-1 genomes and implications for multi-epitope vaccine designs.BMC Microbiol. 2010 Aug 9;10:212. doi: 10.1186/1471-2180-10-212. BMC Microbiol. 2010. PMID: 20696039 Free PMC article.
-
Identification of highly conserved and broadly cross-reactive HIV type 1 cytotoxic T lymphocyte epitopes as candidate immunogens for inclusion in Mycobacterium bovis BCG-vectored HIV vaccines.AIDS Res Hum Retroviruses. 2000 Sep 20;16(14):1433-43. doi: 10.1089/08892220050140982. AIDS Res Hum Retroviruses. 2000. PMID: 11018863 Review.
-
Characteristics of the intrahepatic cytotoxic T lymphocyte response in chronic hepatitis C virus infection.Springer Semin Immunopathol. 1997;19(1):69-83. doi: 10.1007/BF00945026. Springer Semin Immunopathol. 1997. PMID: 9266632 Review.
Cited by
-
Evaluation of the HIV-1 Polymerase Gene Sequence Diversity for Prediction of Recent HIV-1 Infections Using Shannon Entropy Analysis.Viruses. 2022 Jul 21;14(7):1587. doi: 10.3390/v14071587. Viruses. 2022. PMID: 35891568 Free PMC article.
-
Correction to: Deep sequencing of the HIV-1 polymerase gene for characterisation of cytotoxic T-lymphocyte epitopes during early and chronic disease stages.Virol J. 2022 May 5;19(1):78. doi: 10.1186/s12985-022-01803-4. Virol J. 2022. PMID: 35513827 Free PMC article. No abstract available.
References
-
- UNAIDS [Internet]. Fact Sheet 2021 - Preliminary epidemiological estimates. 2021 [accessed July 2021]. Available from: https://www.unaids.org/sites/default/files/media_asset/UNAIDS_FactSheet_....
-
- UNAIDS [Internet]. Global 2019 HIV/AIDS Statistics. 2020 [accessed May 2021]. Available from: https://www.unaids.org/en/resources/documents/2020/unaids-data.
-
- HSRC. The fifth South African national HIV prevalence, incidence, behaviour and communication survey, 2017: HIV impact assessment summary report. HSRC Press Cape Town; 2018.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

