Transcriptome and Small RNA Profiling of Potato Virus Y Infected Potato Cultivars, Including Systemically Infected Russet Burbank
- PMID: 35336930
- PMCID: PMC8952017
- DOI: 10.3390/v14030523
Transcriptome and Small RNA Profiling of Potato Virus Y Infected Potato Cultivars, Including Systemically Infected Russet Burbank
Abstract
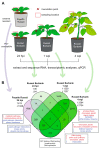
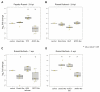
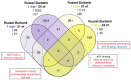
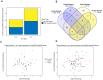
Potatoes are the world's most produced non-grain crops and an important food source for billions of people. Potatoes are susceptible to numerous pathogens that reduce yield, including Potato virus Y (PVY). Genetic resistance to PVY is a sustainable way to limit yield and quality losses due to PVY infection. Potato cultivars vary in their susceptibility to PVY and include susceptible varieties such as Russet Burbank, and resistant varieties such as Payette Russet. Although the loci and genes associated with PVY-resistance have been identified, the genes and mechanisms involved in limiting PVY during the development of systemic infections have yet to be fully elucidated. To increase our understanding of PVY infection, potato antiviral responses, and resistance, we utilized RNA sequencing to characterize the transcriptomes of two potato cultivars. Since transcriptional responses associated with the extreme resistance response occur soon after PVY contact, we analyzed the transcriptome and small RNA profile of both the PVY-resistant Payette Russet cultivar and PVY-susceptible Russet Burbank cultivar 24 hours post-inoculation. While hundreds of genes, including terpene synthase and protein kinase encoding genes, exhibited increased expression, the majority, including numerous genes involved in plant pathogen interactions, were downregulated. To gain greater understanding of the transcriptional changes that occur during the development of systemic PVY-infection, we analyzed Russet Burbank leaf samples one week and four weeks post-inoculation and identified similarities and differences, including higher expression of genes involved in chloroplast function, photosynthesis, and secondary metabolite production, and lower expression of defense response genes at those time points. Small RNA sequencing identified different populations of 21- and 24-nucleotide RNAs and revealed that the miRNA profiles in PVY-infected Russet Burbank plants were similar to those observed in other PVY-tolerant cultivars and that during systemic infection ~32% of the NLR-type disease resistance genes were targeted by 21-nt small RNAs. Analysis of alternative splicing in PVY-infected potato plants identified splice variants of several hundred genes, including isoforms that were more dominant in PVY-infected plants. The description of the PVYN-Wi-associated transcriptome and small RNA profiles of two potato cultivars described herein adds to the body of knowledge regarding differential outcomes of infection for specific PVY strain and host cultivar pairs, which will help further understanding of the mechanisms governing genetic resistance and/or virus-limiting responses in potato plants.
Keywords: Potato virus Y; alternative splicing; extreme resistance; small RNAs; tolerance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
RNA-Seq analysis of resistant and susceptible potato varieties during the early stages of potato virus Y infection.BMC Genomics. 2015 Jun 20;16(1):472. doi: 10.1186/s12864-015-1666-2. BMC Genomics. 2015. PMID: 26091899 Free PMC article.
-
The effects of potato virus Y-derived virus small interfering RNAs of three biologically distinct strains on potato (Solanum tuberosum) transcriptome.Virol J. 2017 Jul 17;14(1):129. doi: 10.1186/s12985-017-0803-8. Virol J. 2017. PMID: 28716126 Free PMC article.
-
Comparative analysis of virus-specific small RNA profiles of three biologically distinct strains of Potato virus Y in infected potato (Solanum tuberosum) cv. Russet Burbank.Virus Res. 2014 Oct 13;191:153-60. doi: 10.1016/j.virusres.2014.07.005. Epub 2014 Jul 15. Virus Res. 2014. PMID: 25036885
-
Insight into aphid mediated Potato Virus Y transmission: A molecular to bioinformatics prospective.Front Microbiol. 2022 Nov 24;13:1001454. doi: 10.3389/fmicb.2022.1001454. eCollection 2022. Front Microbiol. 2022. PMID: 36504828 Free PMC article. Review.
-
Continuous and emerging challenges of Potato virus Y in potato.Annu Rev Phytopathol. 2013;51:571-86. doi: 10.1146/annurev-phyto-082712-102332. Annu Rev Phytopathol. 2013. PMID: 23915135 Review.
Cited by
-
Transcriptomic and functional analyses reveal the molecular mechanisms underlying Fe-mediated tobacco resistance to potato virus Y infection.Front Plant Sci. 2023 Mar 30;14:1163679. doi: 10.3389/fpls.2023.1163679. eCollection 2023. Front Plant Sci. 2023. PMID: 37063211 Free PMC article.
-
Transcriptome analysis of genes involved in the pathogenesis mechanism of potato virus Y in potato cultivar YouJin.Front Microbiol. 2024 Mar 6;15:1353814. doi: 10.3389/fmicb.2024.1353814. eCollection 2024. Front Microbiol. 2024. PMID: 38511006 Free PMC article.
-
Genotyping-by-sequencing and weighted gene co-expression network analysis of genes responsive against Potato virus Y in commercial potato cultivars.PLoS One. 2024 May 24;19(5):e0303783. doi: 10.1371/journal.pone.0303783. eCollection 2024. PLoS One. 2024. PMID: 38787845 Free PMC article.
-
Transcriptional and functional predictors of potato virus Y-induced tuber necrosis in potato (Solanum tuberosum).Front Plant Sci. 2024 Apr 4;15:1369846. doi: 10.3389/fpls.2024.1369846. eCollection 2024. Front Plant Sci. 2024. PMID: 38638354 Free PMC article.
References
-
- Devaux A., Kromann P., Ortiz O. Potatoes for Sustainable Global Food Security. Potato Res. 2014;57:185–199. doi: 10.1007/s11540-014-9265-1. - DOI
-
- Steinwand M.A., Ronald P.C. Crop Biotechnology and the Future of Food. Nat. Food. 2020;1:273–283. doi: 10.1038/s43016-020-0072-3. - DOI
-
- Lal M., Yadav S., Pant R.P., Dua V., Singh B., Kaushik S. Sustainable Potato Production and the Impact of Climate Change. IGI Global; Hershey, PA, USA: 2017. Impact of Global Climate Change on Potato Diseases and Strategies for Their Mitigation; pp. 205–227.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

