Pseudorabies Virus Inhibits Expression of Liver X Receptors to Assist Viral Infection
- PMID: 35336921
- PMCID: PMC8954865
- DOI: 10.3390/v14030514
Pseudorabies Virus Inhibits Expression of Liver X Receptors to Assist Viral Infection
Abstract
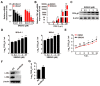
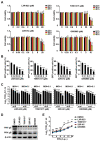
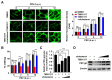
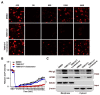
Pseudorabies virus (PRV) is a contagious herpesvirus that causes Aujeszky's disease and economic losses worldwide. Liver X receptors (LXRs) belong to the nuclear receptor superfamily and are critical for the control of lipid homeostasis. However, the role of LXR in PRV infection has not been fully established. In this study, we found that PRV infection downregulated the mRNA and protein levels of LXRα and LXRβ in vitro and in vivo. Furthermore, we discovered that LXR activation suppressed PRV proliferation, while LXR inhibition promoted PRV proliferation. We demonstrated that LXR activation-mediated reduction of cellular cholesterol was critical for the dynamics of PRV entry-dependent clathrin-coated pits. Replenishment of cholesterol restored the dynamics of clathrin-coated pits and PRV entry under LXR activation conditions. Interestingly, T0901317, an LXR agonist, prevented PRV infection in mice. Our results support a model that PRV modulates LXR-regulated cholesterol metabolism to facilitate viral proliferation.
Keywords: Liver X receptors; clathrin-coated pits; pseudorabies virus; viral entry.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


Similar articles
-
Pseudorabies virus upregulates low-density lipoprotein receptors to facilitate viral entry.J Virol. 2024 Jan 23;98(1):e0166423. doi: 10.1128/jvi.01664-23. Epub 2023 Dec 6. J Virol. 2024. PMID: 38054618 Free PMC article.
-
TMEM41B Is an Interferon-Stimulated Gene That Promotes Pseudorabies Virus Replication.J Virol. 2023 Jun 29;97(6):e0041223. doi: 10.1128/jvi.00412-23. Epub 2023 May 31. J Virol. 2023. PMID: 37255475 Free PMC article.
-
Cathelicidin CATH-B1 Inhibits Pseudorabies Virus Infection via Direct Interaction and TLR4/JNK/IRF3-Mediated Interferon Activation.J Virol. 2023 Jul 27;97(7):e0070623. doi: 10.1128/jvi.00706-23. Epub 2023 Jun 14. J Virol. 2023. PMID: 37314341 Free PMC article.
-
Molecular biology of pseudorabies virus: impact on neurovirology and veterinary medicine.Microbiol Mol Biol Rev. 2005 Sep;69(3):462-500. doi: 10.1128/MMBR.69.3.462-500.2005. Microbiol Mol Biol Rev. 2005. PMID: 16148307 Free PMC article. Review.
-
Pseudorabies virus in wild swine: a global perspective.Arch Virol. 2011 Oct;156(10):1691-705. doi: 10.1007/s00705-011-1080-2. Epub 2011 Aug 12. Arch Virol. 2011. PMID: 21837416 Review.
Cited by
-
Pseudorabies virus upregulates low-density lipoprotein receptors to facilitate viral entry.J Virol. 2024 Jan 23;98(1):e0166423. doi: 10.1128/jvi.01664-23. Epub 2023 Dec 6. J Virol. 2024. PMID: 38054618 Free PMC article.
-
Pseudorabies virus manipulates mitochondrial tryptophanyl-tRNA synthetase 2 for viral replication.Virol Sin. 2024 Jun;39(3):403-413. doi: 10.1016/j.virs.2024.04.003. Epub 2024 Apr 16. Virol Sin. 2024. PMID: 38636706 Free PMC article.
-
Reduced NR2F2 Expression in the Host Response to Infectious Bursal Disease Virus Infection Suppressed Viral Replication by Enhancing Type I Interferon Expression by Targeting SOCS5.J Virol. 2023 Jul 27;97(7):e0066423. doi: 10.1128/jvi.00664-23. Epub 2023 Jun 26. J Virol. 2023. PMID: 37358466 Free PMC article.
-
Liver X Receptor-Inducible Host E3 Ligase IDOL Targets a Human Cytomegalovirus Reactivation Determinant.J Virol. 2023 Jul 27;97(7):e0075823. doi: 10.1128/jvi.00758-23. Epub 2023 Jun 20. J Virol. 2023. PMID: 37338407 Free PMC article.
-
Regulation of cholesterol homeostasis in health and diseases: from mechanisms to targeted therapeutics.Signal Transduct Target Ther. 2022 Aug 2;7(1):265. doi: 10.1038/s41392-022-01125-5. Signal Transduct Target Ther. 2022. PMID: 35918332 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

