Insights in Post-Translational Modifications: Ubiquitin and SUMO
- PMID: 35328702
- PMCID: PMC8952880
- DOI: 10.3390/ijms23063281
Insights in Post-Translational Modifications: Ubiquitin and SUMO
Abstract
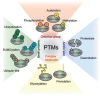
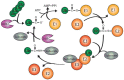
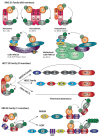
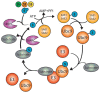
Both ubiquitination and SUMOylation are dynamic post-translational modifications that regulate thousands of target proteins to control virtually every cellular process. Unfortunately, the detailed mechanisms of how all these cellular processes are regulated by both modifications remain unclear. Target proteins can be modified by one or several moieties, giving rise to polymers of different morphology. The conjugation cascades of both modifications comprise a few activating and conjugating enzymes but close to thousands of ligating enzymes (E3s) in the case of ubiquitination. As a result, these E3s give substrate specificity and can form polymers on a target protein. Polymers can be quickly modified forming branches or cleaving chains leading the target protein to its cellular fate. The recent development of mass spectrometry(MS) -based approaches has increased the understanding of ubiquitination and SUMOylation by finding essential modified targets in particular signaling pathways. Here, we perform a concise overview comprising from the basic mechanisms of both ubiquitination and SUMOylation to recent MS-based approaches aimed to find specific targets for particular E3 enzymes.
Keywords: E3 enzymes; SUMO; proteomics; ubiquitin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Molecular mechanisms in SUMO conjugation.Biochem Soc Trans. 2020 Feb 28;48(1):123-135. doi: 10.1042/BST20190357. Biochem Soc Trans. 2020. PMID: 31872228 Review.
-
In Vitro SUMOylation Assay to Study SUMO E3 Ligase Activity.J Vis Exp. 2018 Jan 29;(131):56629. doi: 10.3791/56629. J Vis Exp. 2018. PMID: 29443041 Free PMC article.
-
Structural insights into functional modes of proteins involved in ubiquitin family pathways.Methods Mol Biol. 2012;832:547-76. doi: 10.1007/978-1-61779-474-2_39. Methods Mol Biol. 2012. PMID: 22350912
-
Specific substrate recognition and thioester intermediate determinations in ubiquitin and SUMO conjugation cascades revealed by a high-sensitive FRET assay.Mol Biosyst. 2014 Apr;10(4):778-86. doi: 10.1039/c3mb70155g. Epub 2014 Jan 23. Mol Biosyst. 2014. PMID: 24452848
-
Control of SUMO and Ubiquitin by ROS: Signaling and disease implications.Mol Aspects Med. 2018 Oct;63:3-17. doi: 10.1016/j.mam.2018.07.002. Epub 2018 Aug 4. Mol Aspects Med. 2018. PMID: 30059710 Review.
Cited by
-
BCL-G: 20 years of research on a non-typical protein from the BCL-2 family.Cell Death Differ. 2023 Jun;30(6):1437-1446. doi: 10.1038/s41418-023-01158-5. Epub 2023 Apr 8. Cell Death Differ. 2023. PMID: 37031274 Free PMC article. Review.
-
The role of deubiquitinases in cardiac disease.Expert Rev Mol Med. 2024 Mar 25;26:e3. doi: 10.1017/erm.2024.2. Expert Rev Mol Med. 2024. PMID: 38525836 Free PMC article. Review.
-
Understanding the roles and regulation patterns of circRNA on its host gene in tumorigenesis and tumor progression.J Exp Clin Cancer Res. 2023 Apr 15;42(1):86. doi: 10.1186/s13046-023-02657-6. J Exp Clin Cancer Res. 2023. PMID: 37060016 Free PMC article. Review.
-
SNRPD2 Is a Novel Substrate for the Ubiquitin Ligase Activity of the Salmonella Type III Secretion Effector SlrP.Biology (Basel). 2022 Oct 17;11(10):1517. doi: 10.3390/biology11101517. Biology (Basel). 2022. PMID: 36290420 Free PMC article.
-
Clinical Correlation of Transcription Factor SOX3 in Cancer: Unveiling Its Role in Tumorigenesis.Genes (Basel). 2024 Jun 13;15(6):777. doi: 10.3390/genes15060777. Genes (Basel). 2024. PMID: 38927713 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

