Poxviruses Bearing DNA Polymerase Mutations Show Complex Patterns of Cross-Resistance
- PMID: 35327382
- PMCID: PMC8945813
- DOI: 10.3390/biomedicines10030580
Poxviruses Bearing DNA Polymerase Mutations Show Complex Patterns of Cross-Resistance
Abstract
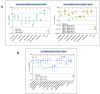
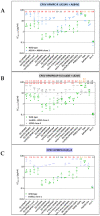
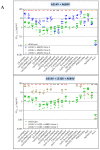
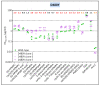
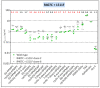
Despite the eradication of smallpox four decades ago, poxviruses continue to be a threat to humans and animals. The arsenal of anti-poxvirus agents is very limited and understanding mechanisms of resistance to agents targeting viral DNA polymerases is fundamental for the development of antiviral therapies. We describe here the phenotypic and genotypic characterization of poxvirus DNA polymerase mutants isolated under selective pressure with different acyclic nucleoside phosphonates, including HPMPC (cidofovir), cHPMPC, HPMPA, cHPMPA, HPMPDAP, HPMPO-DAPy, and PMEO-DAPy, and the pyrophosphate analogue phosphonoacetic acid. Vaccinia virus (VACV) and cowpox virus drug-resistant viral clones emerging under drug pressure were characterized phenotypically (drug-susceptibility profile) and genotypically (DNA polymerase sequencing). Different amino acid changes in the polymerase domain and in the 3'-5' exonuclease domain were linked to drug resistance. Changes in the 3'-5' domain emerged earlier than in the polymerase domain when viruses acquired a combination of mutations. Our study highlights the importance of poxvirus DNA polymerase residues 314, 613, 684, 688, and 851, previously linked to drug resistance, and identified several novel mutations in the 3'-5' exonuclease domain (M313I, F354L, D480Y) and in the DNA polymerase domain (A632T, T831I, E856K, L924F) associated with different drug-susceptibility profiles. Furthermore, a combination of mutations resulted in complex patterns of cross-resistance. Modeling of the VACV DNA polymerase bearing the newly described mutations was performed to understand the effects of these mutations on the structure of the viral enzyme. We demonstrated the emergence of drug-resistant DNA polymerase mutations in complex patterns to be considered in case such mutations should eventually arise in the clinic.
Keywords: DNA polymerase; cidofovir; drug resistance; nucleotide analogues; phosphonoacetic acid; vaccinia virus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures














Similar articles
-
Antiviral potential of a new generation of acyclic nucleoside phosphonates, the 6-[2-(phosphonomethoxy)alkoxy]-2,4-diaminopyrimidines.Nucleosides Nucleotides Nucleic Acids. 2005;24(5-7):331-41. doi: 10.1081/ncn-200059772. Nucleosides Nucleotides Nucleic Acids. 2005. PMID: 16247948
-
Mutations conferring resistance to viral DNA polymerase inhibitors in camelpox virus give different drug-susceptibility profiles in vaccinia virus.J Virol. 2012 Jul;86(13):7310-25. doi: 10.1128/JVI.00355-12. Epub 2012 Apr 24. J Virol. 2012. PMID: 22532673 Free PMC article.
-
DNA polymerase mutations in drug-resistant herpes simplex virus mutants determine in vivo neurovirulence and drug-enzyme interactions.Antivir Ther. 2007;12(5):719-32. Antivir Ther. 2007. PMID: 17713155
-
Vaccinia virus inhibitors as a paradigm for the chemotherapy of poxvirus infections.Clin Microbiol Rev. 2001 Apr;14(2):382-97. doi: 10.1128/CMR.14.2.382-397.2001. Clin Microbiol Rev. 2001. PMID: 11292644 Free PMC article. Review.
-
The acyclic nucleoside phosphonates from inception to clinical use: historical perspective.Antiviral Res. 2007 Jul;75(1):1-13. doi: 10.1016/j.antiviral.2006.10.006. Epub 2006 Nov 7. Antiviral Res. 2007. PMID: 17116336 Review.
Cited by
-
Development of a Novel Loop-Mediated Isothermal Amplification Method for the Rapid Detection of Monkeypox Virus Infections.Viruses. 2022 Dec 28;15(1):84. doi: 10.3390/v15010084. Viruses. 2022. PMID: 36680124 Free PMC article.
-
In silico identification of potential phytochemical inhibitors for mpox virus: molecular docking, MD simulation, and ADMET studies.Mol Divers. 2024 Dec;28(6):4067-4086. doi: 10.1007/s11030-023-10797-2. Epub 2024 Mar 22. Mol Divers. 2024. PMID: 38519803
-
Monkeypox virus emerges from the shadow of its more infamous cousin: family biology matters.Emerg Microbes Infect. 2022 Dec;11(1):1768-1777. doi: 10.1080/22221751.2022.2095309. Emerg Microbes Infect. 2022. PMID: 35751396 Free PMC article. Review.
-
Retrospective Analysis Revealed an April Occurrence of Monkeypox in the Czech Republic.Viruses. 2022 Aug 15;14(8):1773. doi: 10.3390/v14081773. Viruses. 2022. PMID: 36016395 Free PMC article.
-
Human skin-on-a-chip for mpox pathogenesis studies and preclinical drug evaluation.Trends Pharmacol Sci. 2023 Dec;44(12):865-868. doi: 10.1016/j.tips.2023.07.001. Epub 2023 Jul 26. Trends Pharmacol Sci. 2023. PMID: 37500295 Free PMC article.
References
-
- Fenner F. Smallpox: Emergence, global spread, and eradication. Hist. Philos. Life Sci. 1993;15:397–420. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

