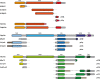
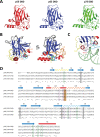
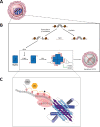
Structural diversity of p63 and p73 isoforms
- PMID: 35314772
- PMCID: PMC9091270
- DOI: 10.1038/s41418-022-00975-4
Structural diversity of p63 and p73 isoforms
Abstract
The p53 protein family is the most studied protein family of all. Sequence analysis and structure determination have revealed a high similarity of crucial domains between p53, p63 and p73. Functional studies, however, have shown a wide variety of different tasks in tumor suppression, quality control and development. Here we review the structure and organization of the individual domains of p63 and p73, the interaction of these domains in the context of full-length proteins and discuss the evolutionary origin of this protein family. FACTS: Distinct physiological roles/functions are performed by specific isoforms. The non-divided transactivation domain of p63 has a constitutively high activity while the transactivation domains of p53/p73 are divided into two subdomains that are regulated by phosphorylation. Mdm2 binds to all three family members but ubiquitinates only p53. TAp63α forms an autoinhibited dimeric state while all other vertebrate p53 family isoforms are constitutively tetrameric. The oligomerization domain of p63 and p73 contain an additional helix that is necessary for stabilizing the tetrameric states. During evolution this helix got lost independently in different phylogenetic branches, while the DNA binding domain became destabilized and the transactivation domain split into two subdomains. OPEN QUESTIONS: Is the autoinhibitory mechanism of mammalian TAp63α conserved in p53 proteins of invertebrates that have the same function of genomic quality control in germ cells? What is the physiological function of the p63/p73 SAM domains? Do the short isoforms of p63 and p73 have physiological functions? What are the roles of the N-terminal elongated TAp63 isoforms, TA* and GTA?
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Regulation of the Activity in the p53 Family Depends on the Organization of the Transactivation Domain.Structure. 2018 Aug 7;26(8):1091-1100.e4. doi: 10.1016/j.str.2018.05.013. Epub 2018 Jun 28. Structure. 2018. PMID: 30099987
-
Identification of DeltaN isoform and polyadenylation site choice variants in molluscan p63/p73-like homologues.Mar Biotechnol (NY). 2007 Mar-Apr;9(2):217-30. doi: 10.1007/s10126-006-6045-1. Epub 2007 Jan 22. Mar Biotechnol (NY). 2007. PMID: 17242983
-
Functional interplay between MDM2, p63/p73 and mutant p53.Oncogene. 2015 Aug 13;34(33):4300-10. doi: 10.1038/onc.2014.359. Epub 2014 Nov 24. Oncogene. 2015. PMID: 25417702 Free PMC article.
-
p53/p63/p73 isoforms: an orchestra of isoforms to harmonise cell differentiation and response to stress.Cell Death Differ. 2006 Jun;13(6):962-72. doi: 10.1038/sj.cdd.4401914. Cell Death Differ. 2006. PMID: 16601753 Review.
-
Evolution of functions within the p53/p63/p73 family.Ann N Y Acad Sci. 2000;926:90-100. doi: 10.1111/j.1749-6632.2000.tb05602.x. Ann N Y Acad Sci. 2000. PMID: 11193045 Review.
Cited by
-
DNp73 enhances tumor progression and immune evasion in multiple myeloma by targeting the MYC and MYCN pathways.Front Immunol. 2024 Sep 24;15:1470328. doi: 10.3389/fimmu.2024.1470328. eCollection 2024. Front Immunol. 2024. PMID: 39380995 Free PMC article.
-
A Rare Case of TP63-Associated Lymphopenia Revealed by Newborn Screening Using TREC.Int J Mol Sci. 2024 Oct 9;25(19):10844. doi: 10.3390/ijms251910844. Int J Mol Sci. 2024. PMID: 39409174 Free PMC article.
-
Disease-related p63 DBD mutations impair DNA binding by distinct mechanisms and varying degree.Cell Death Dis. 2023 Apr 18;14(4):274. doi: 10.1038/s41419-023-05796-y. Cell Death Dis. 2023. PMID: 37072394 Free PMC article.
-
Unlocking the Gateway: The Spatio-Temporal Dynamics of the p53 Family Driven by the Nuclear Pores and Its Implication for the Therapeutic Approach in Cancer.Int J Mol Sci. 2024 Jul 7;25(13):7465. doi: 10.3390/ijms25137465. Int J Mol Sci. 2024. PMID: 39000572 Free PMC article. Review.
-
The Pathology according to p53 Pathway.Pathobiology. 2024;91(3):230-243. doi: 10.1159/000535203. Epub 2023 Nov 14. Pathobiology. 2024. PMID: 37963443 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

