The Evolutionary Advantage in Mammals of the Complementary Monoallelic Expression Mechanism of Genomic Imprinting and Its Emergence From a Defense Against the Insertion Into the Host Genome
- PMID: 35309133
- PMCID: PMC8928582
- DOI: 10.3389/fgene.2022.832983
The Evolutionary Advantage in Mammals of the Complementary Monoallelic Expression Mechanism of Genomic Imprinting and Its Emergence From a Defense Against the Insertion Into the Host Genome
Abstract
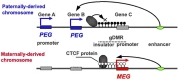
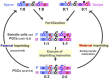
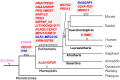
In viviparous mammals, genomic imprinting regulates parent-of-origin-specific monoallelic expression of paternally and maternally expressed imprinted genes (PEGs and MEGs) in a region-specific manner. It plays an essential role in mammalian development: aberrant imprinting regulation causes a variety of developmental defects, including fetal, neonatal, and postnatal lethality as well as growth abnormalities. Mechanistically, PEGs and MEGs are reciprocally regulated by DNA methylation of germ-line differentially methylated regions (gDMRs), thereby exhibiting eliciting complementary expression from parental genomes. The fact that most gDMR sequences are derived from insertion events provides strong support for the claim that genomic imprinting emerged as a host defense mechanism against the insertion in the genome. Recent studies on the molecular mechanisms concerning how the DNA methylation marks on the gDMRs are established in gametes and maintained in the pre- and postimplantation periods have further revealed the close relationship between genomic imprinting and invading DNA, such as retroviruses and LTR retrotransposons. In the presence of gDMRs, the monoallelic expression of PEGs and MEGs confers an apparent advantage by the functional compensation that takes place between the two parental genomes. Thus, it is likely that genomic imprinting is a consequence of an evolutionary trade-off for improved survival. In addition, novel genes were introduced into the mammalian genome via this same surprising and complex process as imprinted genes, such as the genes acquired from retroviruses as well as those that were duplicated by retropositioning. Importantly, these genes play essential/important roles in the current eutherian developmental system, such as that in the placenta and/or brain. Thus, genomic imprinting has played a critically important role in the evolutionary emergence of mammals, not only by providing a means to escape from the adverse effects of invading DNA with sequences corresponding to the gDMRs, but also by the acquisition of novel functions in development, growth and behavior via the mechanism of complementary monoallelic expression.
Keywords: complementation; evolutionary trade-off; genome innovation; host defense hypothesis; insertion of exogenous DNA.
Copyright © 2022 Kaneko-Ishino and Ishino.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Evolution of viviparity in mammals: what genomic imprinting tells us about mammalian placental evolution.Reprod Fertil Dev. 2019 Jul;31(7):1219-1227. doi: 10.1071/RD18127. Reprod Fertil Dev. 2019. PMID: 30625287 Review.
-
The origin and evolution of genomic imprinting and viviparity in mammals.Philos Trans R Soc Lond B Biol Sci. 2013 Jan 5;368(1609):20120151. doi: 10.1098/rstb.2012.0151. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 23166401 Free PMC article. Review.
-
Maternal regulation of chromosomal imprinting in animals.Chromosoma. 2019 Jun;128(2):69-80. doi: 10.1007/s00412-018-00690-5. Epub 2019 Feb 5. Chromosoma. 2019. PMID: 30719566 Free PMC article. Review.
-
The regulation and biological significance of genomic imprinting in mammals.J Biochem. 2003 Jun;133(6):699-711. doi: 10.1093/jb/mvg090. J Biochem. 2003. PMID: 12869525 Review.
-
The evolution of mammalian genomic imprinting was accompanied by the acquisition of novel CpG islands.Genome Biol Evol. 2011;3:1276-83. doi: 10.1093/gbe/evr104. Epub 2011 Oct 19. Genome Biol Evol. 2011. PMID: 22016334 Free PMC article.
Cited by
-
Detection of DNA methylation signatures through the lens of genomic imprinting.Sci Rep. 2024 Jan 19;14(1):1694. doi: 10.1038/s41598-024-52114-3. Sci Rep. 2024. PMID: 38242932 Free PMC article.
-
Advantages of vitrification preservation in assisted reproduction and potential influences on imprinted genes.Clin Epigenetics. 2022 Nov 3;14(1):141. doi: 10.1186/s13148-022-01355-y. Clin Epigenetics. 2022. PMID: 36324168 Free PMC article. Review.
-
PRKACB is a novel imprinted gene in marsupials.Epigenetics Chromatin. 2024 Sep 28;17(1):29. doi: 10.1186/s13072-024-00552-8. Epigenetics Chromatin. 2024. PMID: 39342354 Free PMC article.
-
Endogenous Retroviruses and Placental Evolution, Development, and Diversity.Cells. 2022 Aug 8;11(15):2458. doi: 10.3390/cells11152458. Cells. 2022. PMID: 35954303 Free PMC article. Review.
-
Livestock species as emerging models for genomic imprinting.Front Cell Dev Biol. 2024 Feb 15;12:1348036. doi: 10.3389/fcell.2024.1348036. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38500688 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources

