Unique properties of Coronaviridae single-pass transmembrane domain regions as an adaptation to diverse membrane systems
- PMID: 35306415
- PMCID: PMC8922268
- DOI: 10.1016/j.virol.2022.03.002
Unique properties of Coronaviridae single-pass transmembrane domain regions as an adaptation to diverse membrane systems
Abstract
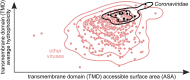
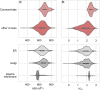
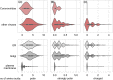
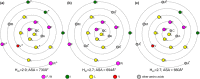
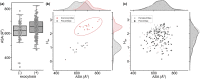
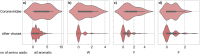
Enveloped viruses such as Coronaviridae (CoV) enter the host cell by fusing the viral envelope directly with the plasma membrane (PM) or with the membrane of the endosome. Replication of the CoV genome takes place in membrane compartments formed by rearrangement of the endoplasmic reticulum (ER) membrane network. Budding of these viruses occurs from the ER-Golgi intermediate compartment (ERGIC). The relationship between proteins and various membranes is crucial for the replication cycle of CoVs. The role of transmembrane domains (TMDs) and pre-transmembrane domains (pre-TMD) of viral proteins in this process is gaining more recognition. Here we present a thorough analysis of physico-chemical parameters, such as accessible surface area (ASA), average hydrophobicity (Hav), and contribution of specific amino acids in TMDs and pre-TMDs of single-span membrane proteins of human viruses. We focus on unique properties of these elements in CoV and postulate their role in adaptation to diverse host membranes and regulation of retention of membrane proteins during replication.
Keywords: Accessible surface area (ASA); Average hydrophobicity (H(av)); Lipid rafts; Retention; Trafficking.
Copyright © 2022 Nencki Institute of Experimental Biology PAS. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
Similar articles
-
Transmembrane Domain Lengths Serve as Signatures of Organismal Complexity and Viral Transport Mechanisms.Sci Rep. 2016 Mar 1;6:22352. doi: 10.1038/srep22352. Sci Rep. 2016. PMID: 26925972 Free PMC article.
-
Type II transmembrane domain hydrophobicity dictates the cotranslational dependence for inversion.Mol Biol Cell. 2014 Nov 1;25(21):3363-74. doi: 10.1091/mbc.E14-04-0874. Epub 2014 Aug 27. Mol Biol Cell. 2014. PMID: 25165139 Free PMC article.
-
Trafficking of tail-anchored proteins: transport from the endoplasmic reticulum to the plasma membrane and sorting between surface domains in polarised epithelial cells.J Cell Sci. 2002 Apr 15;115(Pt 8):1689-702. doi: 10.1242/jcs.115.8.1689. J Cell Sci. 2002. PMID: 11950887
-
Transmembrane Domain Recognition during Membrane Protein Biogenesis and Quality Control.Curr Biol. 2018 Apr 23;28(8):R498-R511. doi: 10.1016/j.cub.2018.02.004. Curr Biol. 2018. PMID: 29689233 Review.
-
Urban planning of the endoplasmic reticulum (ER): how diverse mechanisms segregate the many functions of the ER.Biochim Biophys Acta. 2011 Oct;1813(10):1893-905. doi: 10.1016/j.bbamcr.2011.06.011. Epub 2011 Jul 2. Biochim Biophys Acta. 2011. PMID: 21756943 Free PMC article. Review.
Cited by
-
Viral Membrane Fusion: A Dance Between Proteins and Lipids.Annu Rev Virol. 2023 Sep 29;10(1):139-161. doi: 10.1146/annurev-virology-111821-093413. Annu Rev Virol. 2023. PMID: 37774128 Free PMC article. Review.
References
-
- Bateman A., Martin M.J., Orchard S., Magrane M., Agivetova R., Ahmad S., Alpi E., Bowler-Barnett E.H., Britto R., Bursteinas B., Bye-A-Jee H., Coetzee R., Cukura A., Da Silva A., Denny P., Dogan T., Ebenezer T.G., Fan J., Castro L.G., Garmiri P., Georghiou G., Gonzales L., Hatton-Ellis E., Hussein A., Ignatchenko A., Insana G., Ishtiaq R., Jokinen P., Joshi V., Jyothi D., Lock A., Lopez R., Luciani A., Luo J., Lussi Y., MacDougall A., Madeira F., Mahmoudy M., Menchi M., Mishra A., Moulang K., Nightingale A., Oliveira C.S., Pundir S., Qi G., Raj S., Rice D., Lopez M.R., Saidi R., Sampson J., Sawford T., Speretta E., Turner E., Tyagi N., Vasudev P., Volynkin V., Warner K., Watkins X., Zaru R., Zellner H., Bridge A., Poux S., Redaschi N., Aimo L., Argoud-Puy G., Auchincloss A., Axelsen K., Bansal P., Baratin D., Blatter M.C., Bolleman J., Boutet E., Breuza L., Casals-Casas C., de Castro E., Echioukh K.C., Coudert E., Cuche B., Doche M., Dornevil D., Estreicher A., Famiglietti M.L., Feuermann M., Gasteiger E., Gehant S., Gerritsen V., Gos A., Gruaz-Gumowski N., Hinz U., Hulo C., Hyka-Nouspikel N., Jungo F., Keller G., Kerhornou A., Lara V., Le Mercier P., Lieberherr D., Lombardot T., Martin X., Masson P., Morgat A., Neto T.B., Paesano S., Pedruzzi I., Pilbout S., Pourcel L., Pozzato M., Pruess M., Rivoire C., Sigrist C., Sonesson K., Stutz A., Sundaram S., Tognolli M., Verbregue L., Wu C.H., Arighi C.N., Arminski L., Chen C., Chen Y., Garavelli J.S., Huang H., Laiho K., McGarvey P., Natale D.A., Ross K., Vinayaka C.R., Wang Q., Wang Y., Yeh L.S., Zhang J. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–D489. doi: 10.1093/nar/gkaa1100. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

