Characterizing kinase intergenic-breakpoint rearrangements in a large-scale lung cancer population and real-world clinical outcomes
- PMID: 35305401
- PMCID: PMC9058911
- DOI: 10.1016/j.esmoop.2022.100405
Characterizing kinase intergenic-breakpoint rearrangements in a large-scale lung cancer population and real-world clinical outcomes
Abstract
Background: Kinase gene fusions are strong driver mutations in neoplasia; however, kinase intergenic-breakpoint rearrangements (IGRs) confound the detection of such fusions and of targeted treatments. We aim to provide an overview of kinase IGRs in a large lung cancer cohort and examine real-world survival outcomes of patients with such fusions.
Methods: Mutational profiles analyzed using targeted next-generation sequencing of 425 cancer-related genes between June 2016 and July 2019 were retrospectively reviewed. Patients' demographic data, clinical characteristics, and survivals were analyzed. RNA sequencing or immunohistochemical assays were carried out to verify chimeric fusion products.
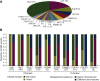
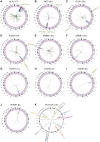
Results: We identified 3411 patients with kinase fusions from a cohort of 30 450 patients with lung cancer, and 624 kinase IGR events were identified in 538 of the 3411 patients. The most frequently identified kinase genes included anaplastic lymphoma kinase (ALK), RET proto-oncogene (RET), ROS proto-oncogene 1 (ROS1), Erb-B2 receptor tyrosine kinase 2/3 (ERBB2/3), and epidermal growth factor receptor (EGFR). Our data showed that most (67%) kinase IGRs occurred on the same chromosome and kinase domains remained intact at the 3'-end. Approximately 3% (19/624) of the kinase IGRs had one genomic breakpoint located in gene promoter regions, including nine fusion events involving ALK, RET, ROS1, EGFR, ERBB2, or fibroblast growth factor receptor 3 (FGFR3). Among the 538 patients with kinase IGRs, 167 (31%) lacked oncogenic driver mutations, among which 28 received targeted therapies in real-world practice. Notably, three ALK IGR patients who harbored no canonical oncogenic aberrations were confirmed with an EML4-ALK chimeric fusion product by RNA sequencing and/or ALK immunohistochemical assays. One patient demonstrated a favorable clinical outcome after 14 months on crizotinib. An additional two patients who had ROS1 IGRs demonstrated a clinical benefit after 13 and 19 months on crizotinib, respectively.
Conclusion: A large real-world lung cancer cohort with kinase IGRs was comprehensively analyzed for their molecular characteristics. The data indicated the potential oncogenic function of kinase IGRs and their outcomes following the administration of targeted therapies.
Keywords: intergenic breakpoints; kinase fusion; lung cancer; real-world survival; targeted next-generation sequencing.
Copyright © 2022 The Authors. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Disclosure YM, QO, and XW are employees of Nanjing Geneseeq Technology Inc., Jiangsu, China. All other authors have declared no conflicts of interest.
Figures


Similar articles
-
Potential Unreliability of Uncommon ALK, ROS1, and RET Genomic Breakpoints in Predicting the Efficacy of Targeted Therapy in NSCLC.J Thorac Oncol. 2021 Mar;16(3):404-418. doi: 10.1016/j.jtho.2020.10.156. Epub 2020 Nov 26. J Thorac Oncol. 2021. PMID: 33248323
-
Intergenic Breakpoints Identified by DNA Sequencing Confound Targetable Kinase Fusion Detection in NSCLC.J Thorac Oncol. 2020 Jul;15(7):1223-1231. doi: 10.1016/j.jtho.2020.02.023. Epub 2020 Mar 7. J Thorac Oncol. 2020. PMID: 32151779
-
Management of advanced non-small cell lung cancers with known mutations or rearrangements: latest evidence and treatment approaches.Ther Adv Respir Dis. 2016 Apr;10(2):113-29. doi: 10.1177/1753465815617871. Epub 2015 Nov 30. Ther Adv Respir Dis. 2016. PMID: 26620497 Free PMC article. Review.
-
Targeted next-generation-sequencing for reliable detection of targetable rearrangements in lung adenocarcinoma-a single center retrospective study.Pathol Res Pract. 2018 Apr;214(4):572-578. doi: 10.1016/j.prp.2018.02.001. Epub 2018 Feb 16. Pathol Res Pract. 2018. PMID: 29580750 Free PMC article.
-
Targeted therapies in non-small cell lung cancer: a focus on ALK/ROS1 tyrosine kinase inhibitors.Expert Rev Anticancer Ther. 2018 Jan;18(1):71-80. doi: 10.1080/14737140.2018.1412260. Epub 2017 Dec 6. Expert Rev Anticancer Ther. 2018. PMID: 29187012 Review.
Cited by
-
Superior clinical outcomes in patients with non-small cell lung cancer harboring multiple ALK fusions treated with tyrosine kinase inhibitors.Transl Lung Cancer Res. 2023 Sep 28;12(9):1935-1948. doi: 10.21037/tlcr-23-484. Epub 2023 Sep 18. Transl Lung Cancer Res. 2023. PMID: 37854161 Free PMC article.
-
Landscape of potentially targetable receptor tyrosine kinase fusions in diverse cancers by DNA-based profiling.NPJ Precis Oncol. 2022 Nov 11;6(1):84. doi: 10.1038/s41698-022-00325-0. NPJ Precis Oncol. 2022. PMID: 36369474 Free PMC article.
-
Pan-cancer molecular analysis of EGFR large fragment deletion in the Asian population.Cancer Med. 2023 Apr;12(7):8083-8088. doi: 10.1002/cam4.5603. Epub 2023 Jan 9. Cancer Med. 2023. PMID: 36622089 Free PMC article.
References
-
- Soda M., Choi Y.L., Enomoto M., et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561–566. - PubMed
-
- Wang R., Hu H., Pan Y., et al. RET fusions define a unique molecular and clinicopathologic subtype of non-small-cell lung cancer. J Clin Oncol. 2012;30(35):4352–4359. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

