Src acts with WNT/FGFRL signaling to pattern the planarian anteroposterior axis
- PMID: 35297964
- PMCID: PMC8995084
- DOI: 10.1242/dev.200125
Src acts with WNT/FGFRL signaling to pattern the planarian anteroposterior axis
Abstract
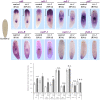
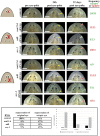
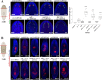
Tissue identity determination is crucial for regeneration, and the planarian anteroposterior (AP) axis uses positional control genes expressed from body wall muscle to determine body regionalization. Canonical Wnt signaling establishes anterior versus posterior pole identities through notum and wnt1 signaling, and two Wnt/FGFRL signaling pathways control head and trunk domains, but their downstream signaling mechanisms are not fully understood. Here, we identify a planarian Src homolog that restricts head and trunk identities to anterior positions. src-1(RNAi) animals formed enlarged brains and ectopic eyes and also duplicated trunk tissue, similar to a combination of Wnt/FGFRL RNAi phenotypes. src-1 was required for establishing territories of positional control gene expression in Schmidtea mediterranea, indicating that it acts at an upstream step in patterning the AP axis. Double RNAi experiments and eye regeneration assays suggest src-1 can act in parallel to at least some Wnt and FGFRL factors. Co-inhibition of src-1 with other posterior-promoting factors led to dramatic patterning changes and a reprogramming of Wnt/FGFRLs into controlling new positional outputs. These results identify src-1 as a factor that promotes robustness of the AP positional system that instructs appropriate regeneration.
Keywords: Axis formation; Patterning; Planarian; Positional information; Regeneration; Src; Wnt.
© 2022. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures



Similar articles
-
Two FGFRL-Wnt circuits organize the planarian anteroposterior axis.Elife. 2016 Apr 11;5:e12845. doi: 10.7554/eLife.12845. Elife. 2016. PMID: 27063937 Free PMC article.
-
Wnt, Ptk7, and FGFRL expression gradients control trunk positional identity in planarian regeneration.Elife. 2016 Apr 13;5:e12850. doi: 10.7554/eLife.12850. Elife. 2016. PMID: 27074666 Free PMC article.
-
BMP suppresses WNT to integrate patterning of orthogonal body axes in adult planarians.PLoS Genet. 2023 Sep 20;19(9):e1010608. doi: 10.1371/journal.pgen.1010608. eCollection 2023 Sep. PLoS Genet. 2023. PMID: 37729232 Free PMC article.
-
Signaling pathways and axis formation in the lower metazoa.Curr Top Dev Biol. 2011;97:137-77. doi: 10.1016/B978-0-12-385975-4.00012-7. Curr Top Dev Biol. 2011. PMID: 22074605 Review.
-
Intercalary regeneration in planarians.Dev Dyn. 2003 Feb;226(2):308-16. doi: 10.1002/dvdy.10249. Dev Dyn. 2003. PMID: 12557208 Review.
Cited by
-
Cellular diversity and developmental hierarchy in the planarian nervous system.Curr Opin Genet Dev. 2022 Oct;76:101960. doi: 10.1016/j.gde.2022.101960. Epub 2022 Jul 22. Curr Opin Genet Dev. 2022. PMID: 35878572 Free PMC article. Review.
-
Djck1α Is Required for Proper Regeneration and Maintenance of the Medial Tissues in Planarians.Cells. 2023 Feb 1;12(3):473. doi: 10.3390/cells12030473. Cells. 2023. PMID: 36766815 Free PMC article.
-
Spatiotemporal transcriptomic atlas reveals the dynamic characteristics and key regulators of planarian regeneration.Nat Commun. 2023 Jun 2;14(1):3205. doi: 10.1038/s41467-023-39016-0. Nat Commun. 2023. PMID: 37268637 Free PMC article.
-
map3k1 suppresses terminal differentiation of migratory eye progenitors in planarian regeneration.bioRxiv [Preprint]. 2024 Oct 12:2024.10.11.617745. doi: 10.1101/2024.10.11.617745. bioRxiv. 2024. PMID: 39416008 Free PMC article. Preprint.
-
Mechanistic regulation of planarian shape during growth and degrowth.Development. 2024 May 1;151(9):dev202353. doi: 10.1242/dev.202353. Epub 2024 May 9. Development. 2024. PMID: 38619319
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

