Identification of Novel 5' and 3' Translation Enhancers in Umbravirus-Like Coat Protein-Deficient RNA Replicons
- PMID: 35297668
- PMCID: PMC9006961
- DOI: 10.1128/jvi.01736-21
Identification of Novel 5' and 3' Translation Enhancers in Umbravirus-Like Coat Protein-Deficient RNA Replicons
Abstract
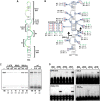
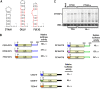
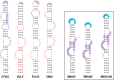
Translation of plant plus-strand RNA viral genomes that lack a 5' cap frequently requires the use of cap-independent translation enhancers (CITEs) located in or near the 3' untranslated region (UTR). 3'CITEs are grouped based on secondary structure and ability to interact with different translation initiation factors or ribosomal subunits, which assemble a complex at the 3' end that is nearly always transferred to the 5' end via a long-distance kissing-loop interaction between sequences in the 3'CITE and 5' hairpins. We report here the identification of a novel 3'CITE in coat protein-deficient RNA replicons that are related to umbraviruses. Umbra-like associated RNAs (ulaRNAs), such as citrus yellow vein-associated virus (CYVaV), are a new type of subviral RNA that do not encode movement proteins, coat proteins, or silencing suppressors but can independently replicate using their encoded RNA-dependent RNA polymerase. An extended hairpin structure containing multiple internal loops in the 3' UTR of CYVaV is strongly conserved in the most closely related ulaRNAs and structurally resembles an I-shaped structure (ISS) 3'CITE. However, unlike ISS, the CYVaV structure binds to eIF4G and no long-distance interaction is discernible between the CYVaV ISS-like structure and sequences at or near the 5' end. We also report that the ∼30-nucleotide (nt) 5' terminal hairpin of CYVaV and related ulaRNAs can enhance translation of reporter constructs when associated with either the CYVaV 3'CITE or the 3'CITEs of umbravirus pea enation mosaic virus (PEMV2) and even independent of a 3'CITE. These findings introduce a new type of 3'CITE and provide the first information on translation of ulaRNAs. IMPORTANCE Umbra-like associated RNAs (ulaRNAs) are a recently discovered type of subviral RNA that use their encoded RNA-dependent RNA polymerase for replication but do not encode any coat proteins, movement proteins, or silencing suppressors yet can be found in plants in the absence of any discernible helper virus. We report the first analysis of their translation using class 2 ulaRNA citrus yellow vein-associated virus (CYVaV). CYVaV uses a novel eIF4G-binding I-shaped structure as its 3' cap-independent translation enhancer (3'CITE), which does not connect with the 5' end by a long-distance RNA:RNA interaction that is typical of 3'CITEs. ulaRNA 5' terminal hairpins can also enhance translation in association with cognate 3'CITEs or those of related ulaRNAs and, to a lesser extent, with 3'CITEs of umbraviruses, or even independent of a 3'CITE. These findings introduce a new type of 3'CITE and provide the first information on translation of ulaRNAs.
Keywords: 3′CITEs; RNA structure; eIF4G-binding structure; noncanonical translation; translation enhancer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
The 3' untranslated region of Pea Enation Mosaic Virus contains two T-shaped, ribosome-binding, cap-independent translation enhancers.J Virol. 2014 Oct;88(20):11696-712. doi: 10.1128/JVI.01433-14. Epub 2014 Aug 6. J Virol. 2014. PMID: 25100834 Free PMC article.
-
Conserved Structure Associated with Different 3'CITEs Is Important for Translation of Umbraviruses.Viruses. 2023 Feb 27;15(3):638. doi: 10.3390/v15030638. Viruses. 2023. PMID: 36992347 Free PMC article.
-
Structural Analysis and Whole Genome Mapping of a New Type of Plant Virus Subviral RNA: Umbravirus-Like Associated RNAs.Viruses. 2021 Apr 9;13(4):646. doi: 10.3390/v13040646. Viruses. 2021. PMID: 33918656 Free PMC article.
-
3' Cap-independent translation enhancers of positive-strand RNA plant viruses.Curr Opin Virol. 2011 Nov;1(5):373-80. doi: 10.1016/j.coviro.2011.10.002. Epub 2011 Oct 24. Curr Opin Virol. 2011. PMID: 22440838 Review.
-
3'UTRs of carmoviruses.Virus Res. 2015 Aug 3;206:27-36. doi: 10.1016/j.virusres.2015.01.023. Epub 2015 Feb 4. Virus Res. 2015. PMID: 25662021 Review.
Cited by
-
Metagenomic identification of novel viruses of maize and teosinte in North America.BMC Genomics. 2022 Nov 23;23(1):767. doi: 10.1186/s12864-022-09001-w. BMC Genomics. 2022. PMID: 36418948 Free PMC article.
-
Grapevine Virome of the Don Ampelographic Collection in Russia Has Concealed Five Novel Viruses.Viruses. 2023 Dec 14;15(12):2429. doi: 10.3390/v15122429. Viruses. 2023. PMID: 38140672 Free PMC article.
-
Direct nanopore RNA sequencing of umbra-like virus-infected plants reveals long non-coding RNAs, specific cleavage sites, D-RNAs, foldback RNAs, and temporal- and tissue-specific profiles.NAR Genom Bioinform. 2024 Aug 16;6(3):lqae104. doi: 10.1093/nargab/lqae104. eCollection 2024 Sep. NAR Genom Bioinform. 2024. PMID: 39157584 Free PMC article.
-
Molecular characterization of a novel umbra-like virus from Thuja orientalis (arborvitae) in South Korea.Arch Virol. 2023 Jul 1;168(7):197. doi: 10.1007/s00705-023-05783-w. Arch Virol. 2023. PMID: 37392254
-
Two new umbravirus-like associated RNAs (ulaRNAs) discovered in maize and johnsongrass from Ecuador.Arch Virol. 2022 Oct;167(10):2093-2098. doi: 10.1007/s00705-022-05525-4. Epub 2022 Jul 12. Arch Virol. 2022. PMID: 35821148
References
-
- Pestova TV, Lorsch JR, Hellen CUT. 2007. The mechanism of translation initiation in eukaryotes, p 87–128. In Mathews MB, Sonenberg N, Hershey JWB (ed), Translational control in biology and medicine. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

