Targeted long-read sequencing reveals clonally expanded HBV-associated chromosomal translocations in patients with chronic hepatitis B
- PMID: 35295767
- PMCID: PMC8918852
- DOI: 10.1016/j.jhepr.2022.100449
Targeted long-read sequencing reveals clonally expanded HBV-associated chromosomal translocations in patients with chronic hepatitis B
Abstract
Background & aims: HBV infects over 257 million people worldwide and is associated with the development of hepatocellular carcinoma (HCC). Integration of HBV DNA into the host genome is likely a key driver of HCC oncogenesis. Here, we utilise targeted long-read sequencing to determine the structure of HBV DNA integrations as well as full isoform information of HBV mRNA with more accurate quantification than traditional next generation sequencing platforms.
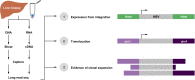
Methods: DNA and RNA were isolated from fresh frozen liver biopsies collected within the GS-US-174-0149 clinical trial. A pan-genotypic panel of biotinylated oligos was developed to enrich for HBV sequences from sheared genomic DNA (∼7 kb) and full-length cDNA libraries from poly-adenylated RNA. Samples were sequenced on the PacBio long-read platform and analysed using a custom bioinformatic pipeline.
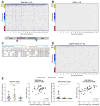
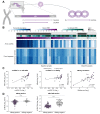
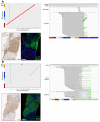
Results: HBV-targeted long-read DNA sequencing generated high coverage data spanning entire integrations. Strikingly, in 13 of 42 samples (31%) we were able to detect HBV sequences flanked by 2 different chromosomes, indicating a chromosomal translocation associated with HBV integration. Chromosomal translocations were unique to each biopsy sample, suggesting that each originated randomly, and in some cases had evidence of clonal expansion. Using targeted long-read RNA sequencing, we determined that upwards of 95% of all HBV transcripts in patients who are HBeAg-positive originate from cccDNA. In contrast, patients who are HBeAg-negative expressed mostly HBsAg from integrations.
Conclusions: Targeted lso-Seq allowed for accurate quantitation of the HBV transcriptome and assignment of transcripts to either cccDNA or integration origins. The existence of multiple unique HBV-associated inter-chromosomal translocations in non-HCC CHB patient liver biopsies suggests a novel mechanism with mutagenic potential that may contribute to progression to HCC.
Lay summary: Fresh frozen liver biopsies from patients infected with HBV were subjected to targeted long-read RNA and DNA sequencing. Long-read RNA sequencing captures entire HBV transcripts in a single read, allowing for resolution of overlapping transcripts from the HBV genome. This resolution allowed us to quantify the burden of transcription from integrations vs. cccDNA origin in individual patients. Patients who were HBeAg-positive had a significantly larger fraction of the HBV transcriptome originating from cccDNA compared with those who were HBeAg-negative. Long-read DNA sequencing captured entire integrated HBV sequences including multiple kilobases of flanking host sequence within single reads. This resolution allowed us to describe integration events flanked by 2 different host chromosomes, indicating that integrated HBV DNA are associated with inter-chromosomal translocations. This may lead to significant transcriptional dysregulation and drive progression to HCC.
Keywords: CCS, circular consensus sequence; CHB, chronic hepatitis B; Chromosomal translocations; Chronic HBV; Clonal expansion; DNA-Seq, DNA sequencing; DR1, direct repeat 1; FFPE, formalin-fixed paraffin-embedded; HCC, hepatocellular carcinoma; Integrated HBV DNA; Long-read sequencing; N/A, nucleos(t)ide analogue; NHEJ, non-homologous end-joining; PEG-IFNα, pegylated interferon α; RIN, RNA integrity number; RNA-Seq, RNA sequencing; TDF, tenofovir disoproxil fumarate; TERT, telomerase reverse transcriptase; Targeted sequencing; WGS, whole genome sequencing; cccDNA, covalently closed circular DNA; contig, contiguous sequence; dslDNA, double-stranded linear DNA; gDNA, genomic DNA; pgRNA, pre-genomic RNA; rcDNA, relaxed circular DNA; targeted Iso-Seq, targeted long-read RNA-sequencing.
© 2022 The Authors.
Conflict of interest statement
All authors are employed by Gilead Sciences Inc. Please refer to the accompanying ICMJE disclosure forms for further details.
Figures


Similar articles
-
Targeted Long-Read Sequencing Reveals Comprehensive Architecture, Burden, and Transcriptional Signatures from Hepatitis B Virus-Associated Integrations and Translocations in Hepatocellular Carcinoma Cell Lines.J Virol. 2021 Sep 9;95(19):e0029921. doi: 10.1128/JVI.00299-21. Epub 2021 Jul 21. J Virol. 2021. PMID: 34287049 Free PMC article.
-
Intrahepatic quantification of HBV antigens in chronic hepatitis B reveals heterogeneity and treatment-mediated reductions in HBV core-positive cells.JHEP Rep. 2022 Dec 26;5(4):100664. doi: 10.1016/j.jhepr.2022.100664. eCollection 2023 Apr. JHEP Rep. 2022. PMID: 36908748 Free PMC article.
-
Reduction of covalently closed circular DNA with long-term nucleos(t)ide analogue treatment in chronic hepatitis B.J Hepatol. 2017 Feb;66(2):275-281. doi: 10.1016/j.jhep.2016.08.022. Epub 2016 Sep 14. J Hepatol. 2017. PMID: 27639844
-
The roadmap towards cure of chronic hepatitis B virus infection.J R Soc N Z. 2020 Oct 15;52(2):129-148. doi: 10.1080/03036758.2020.1811355. eCollection 2022. J R Soc N Z. 2020. PMID: 39439817 Free PMC article. Review.
-
Progression and status of antiviral monitoring in patients with chronic hepatitis B: From HBsAg to HBV RNA.World J Hepatol. 2018 Sep 27;10(9):603-611. doi: 10.4254/wjh.v10.i9.603. World J Hepatol. 2018. PMID: 30310538 Free PMC article. Review.
Cited by
-
Integrated hepatitis B virus DNA maintains surface antigen production during antiviral treatment.J Clin Invest. 2022 Sep 15;132(18):e161818. doi: 10.1172/JCI161818. J Clin Invest. 2022. PMID: 35797115 Free PMC article.
-
Hepatitis B virus enhancer 1 activates preS1 and preS2 promoters of integrated HBV DNA impairing HBsAg secretion.JHEP Rep. 2024 Jun 13;6(9):101144. doi: 10.1016/j.jhepr.2024.101144. eCollection 2024 Sep. JHEP Rep. 2024. PMID: 39253701 Free PMC article.
-
Efficacy of antiviral therapy and host-virus interactions visualised using serial liver sampling with fine-needle aspirates.JHEP Rep. 2023 Jun 19;5(9):100817. doi: 10.1016/j.jhepr.2023.100817. eCollection 2023 Sep. JHEP Rep. 2023. PMID: 37600958 Free PMC article.
-
HBV eradication from the host: Current understanding and challenges.Clin Liver Dis (Hoboken). 2024 Jun 5;23(1):e0188. doi: 10.1097/CLD.0000000000000188. eCollection 2024 Jan-Jun. Clin Liver Dis (Hoboken). 2024. PMID: 38841198 Free PMC article. Review.
-
An enrichment protocol and analysis pipeline for long read sequencing of the hepatitis B virus transcriptome.J Gen Virol. 2023 May;104(5):001856. doi: 10.1099/jgv.0.001856. J Gen Virol. 2023. PMID: 37196057 Free PMC article.
References
-
- World Health Organization . 2017. Hepatitis B.https://www.who.int/news-room/fact-sheets/detail/hepatitis-b
-
- Toh S.T., Jin Y., Liu L., Wang J., Babrzadeh F., Gharizadeh B., et al. Deep sequencing of the hepatitis B virus in hepatocellular carcinoma patients reveals enriched integration events, structural alterations and sequence variations. Carcinogenesis. 2013;34:787–798. - PubMed
-
- Lau C.C., Sun T., Ching A.K., He M., Li J.W., Wong A.M., et al. Viral-human chimeric transcript predisposes risk to liver cancer development and progression. Cancer Cell. 2014;25:335–349. - PubMed
LinkOut - more resources
Full Text Sources

