Decoding the Role of Astrocytes in the Entorhinal Cortex in Alzheimer's Disease Using High-Dimensional Single-Nucleus RNA Sequencing Data and Next-Generation Knowledge Discovery Methodologies: Focus on Drugs and Natural Product Remedies for Dementia
- PMID: 35295737
- PMCID: PMC8918735
- DOI: 10.3389/fphar.2021.720170
Decoding the Role of Astrocytes in the Entorhinal Cortex in Alzheimer's Disease Using High-Dimensional Single-Nucleus RNA Sequencing Data and Next-Generation Knowledge Discovery Methodologies: Focus on Drugs and Natural Product Remedies for Dementia
Abstract
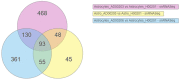
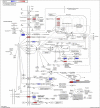
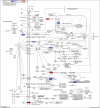
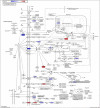
Introduction: Alzheimer's disease (AD) is a major cause of the development of cognitive decline and dementia. AD and associated dementias (ADRD) are the major contributors to the enormous burden of morbidity and mortality worldwide. To date, there are no robust therapies to alleviate or cure this debilitating disease. Most drug treatments focus on restoring the normal function of neurons and the cells that cause inflammation, such as microglia in the brain. However, the role of astrocytes, the brain's housekeeping cells, in the development of AD and the initiation of dementia is still not well understood. Objective: To decipher the role of astrocytes in the entorhinal cortex of AD patients using single nuclear RNA sequencing (snRNASeq) datasets from the Single Cell RNA-seq Database for Alzheimer's Disease (scREAD). The datasets were originally derived from astrocytes, isolated from the entorhinal cortex of AD brain and healthy brain to decipher disease-specific signaling pathways as well as drugs and natural products that reverse AD-specific signatures in astrocytes. Methods: We used snRNASeq datasets from the scREAD database originally derived from astrocytes isolated from the entorhinal cortex of AD and healthy brains from the Gene Expression Omnibus (GEO) (GSE138852 and GSE147528) and analyzed them using next-generation knowledge discovery (NGKD) platforms. scREAD is a user-friendly open-source interface available at https://bmbls.bmi.osumc.edu/scread/that enables more discovery-oriented strategies. snRNASeq data and metadata can also be visualized and downloaded via an interactive web application at adsn.ddnetbio.com. Differentially expressed genes (DEGs) for each snRNASeq dataset were analyzed using iPathwayGuide to compare and derive disease-specific pathways, gene ontologies, and in silico predictions of drugs and natural products that regulate AD -specific signatures in astrocytes. In addition, DEGs were analyzed using the L1000FWD and L1000CDS2 signature search programming interfaces (APIs) to identify additional drugs and natural products that mimic or reverse AD-specific gene signatures in astrocytes. Results: We found that PI3K/AKT signaling, Wnt signaling, neuroactive ligand-receptor interaction pathways, neurodegeneration pathways, etc. were significantly impaired in astrocytes from the entorhinal cortex of AD patients. Biological processes such as glutamate receptor signaling pathway, regulation of synapse organization, cell-cell adhesion via plasma membrane adhesion molecules, and chylomicrons were negatively enriched in the astrocytes from the entorhinal cortex of AD patients. Gene sets involved in cellular components such as postsynaptic membrane, synaptic membrane, postsynapse, and synapse part were negatively enriched (p < 0.01). Moreover, molecular functions such as glutamate receptor activity, neurotransmitter receptor activity, and extracellular ligand-gated ion channels were negatively regulated in the astrocytes of the entorhinal cortex of AD patients (p < 0.01). Moreover, the application of NGKD platforms revealed that antirheumatic drugs, vitamin-E, emetine, narciclasine, cephaeline, trichostatin A, withaferin A, dasatinib, etc. can potentially reverse gene signatures associated with AD. Conclusions: The present study highlights an innovative approach to use NGKD platforms to find unique disease-associated signaling pathways and specific synthetic drugs and natural products that can potentially reverse AD and ADRD-associated gene signatures.
Keywords: alzheimer’s disease and dementia; anti-rheumatic agents; astrocytes; dasatinib; in silico tools; natural products; scREAD; single-nucleus RNA sequencing.
Copyright © 2022 Pushparaj, Kalamegam, Wali Sait and Rasool.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Deciphering aging-associated molecular mechanisms in bone marrow derived hematopoietic stem cells in the elderly using NGS data.Bioinformation. 2024 Feb 29;20(2):180-189. doi: 10.6026/973206300200180. eCollection 2024. Bioinformation. 2024. PMID: 38497076 Free PMC article.
-
Identification of Novel Gene Signatures using Next-Generation Sequencing Data from COVID-19 Infection Models: Focus on Neuro-COVID and Potential Therapeutics.Front Pharmacol. 2021 Aug 31;12:688227. doi: 10.3389/fphar.2021.688227. eCollection 2021. Front Pharmacol. 2021. PMID: 34531741 Free PMC article.
-
FTH1- and SAT1-Induced Astrocytic Ferroptosis Is Involved in Alzheimer's Disease: Evidence from Single-Cell Transcriptomic Analysis.Pharmaceuticals (Basel). 2022 Sep 22;15(10):1177. doi: 10.3390/ph15101177. Pharmaceuticals (Basel). 2022. PMID: 36297287 Free PMC article.
-
Revealing cell vulnerability in Alzheimer's disease by single-cell transcriptomics.Semin Cell Dev Biol. 2023 Apr;139:73-83. doi: 10.1016/j.semcdb.2022.05.007. Epub 2022 May 24. Semin Cell Dev Biol. 2023. PMID: 35623983 Review.
-
Therapeutic potentials of plant iridoids in Alzheimer's and Parkinson's diseases: A review.Eur J Med Chem. 2019 May 1;169:185-199. doi: 10.1016/j.ejmech.2019.03.009. Epub 2019 Mar 8. Eur J Med Chem. 2019. PMID: 30877973 Review.
Cited by
-
Alterations of mRNAs and Non-coding RNAs Associated with Neuroinflammation in Alzheimer's Disease.Mol Neurobiol. 2024 Aug;61(8):5826-5840. doi: 10.1007/s12035-023-03908-5. Epub 2024 Jan 18. Mol Neurobiol. 2024. PMID: 38236345 Review.
-
Role of the antineoplastic drug bleomycin based on toxicogenomic-DNA damage inducing (TGx-DDI) genomic biomarkers data: A meta-analysis.Pak J Med Sci. 2023 Mar-Apr;39(2):423-429. doi: 10.12669/pjms.39.2.7321. Pak J Med Sci. 2023. PMID: 36950431 Free PMC article.
-
Deciphering aging-associated molecular mechanisms in bone marrow derived hematopoietic stem cells in the elderly using NGS data.Bioinformation. 2024 Feb 29;20(2):180-189. doi: 10.6026/973206300200180. eCollection 2024. Bioinformation. 2024. PMID: 38497076 Free PMC article.
-
Exploring the multifaceted therapeutic mechanism of Schisanlactone E (XTS) in APP/PS1 mouse model of Alzheimer's disease through multi-omics analysis.Front Microbiol. 2024 Jul 9;15:1440564. doi: 10.3389/fmicb.2024.1440564. eCollection 2024. Front Microbiol. 2024. PMID: 39044957 Free PMC article.
-
Integrative multi-omics and systems bioinformatics in translational neuroscience: A data mining perspective.J Pharm Anal. 2023 Aug;13(8):836-850. doi: 10.1016/j.jpha.2023.06.011. Epub 2023 Jun 30. J Pharm Anal. 2023. PMID: 37719197 Free PMC article. Review.
References
-
- Ahmad S. S., Khan H., Danish Rizvi S. M., Ansari S. A., Ullah R., Rastrelli L., et al. (2019). Computational Study of Natural Compounds for the Clearance of Amyloid-Βeta: A Potential Therapeutic Management Strategy for Alzheimer's Disease. Molecules 24 (18), 3233. 10.3390/molecules24183233 - DOI - PMC - PubMed
-
- Bahlas S., Damiati L., Dandachi N., Sait H., Alsefri M., Pushparaj P. N. (2019). Rapid Immunoprofiling of Cytokines, Chemokines and Growth Factors in Patients with Active Rheumatoid Arthritis Using Luminex Multiple Analyte Profiling Technology for Precision Medicine. Clin. Exp. Rheumatol. 37 (1), 112–119. - PubMed
LinkOut - more resources
Full Text Sources

