Cytosolic O-GlcNAcylation and PNG1 maintain Drosophila gut homeostasis by regulating proliferation and apoptosis
- PMID: 35294432
- PMCID: PMC8959174
- DOI: 10.1371/journal.pgen.1010128
Cytosolic O-GlcNAcylation and PNG1 maintain Drosophila gut homeostasis by regulating proliferation and apoptosis
Abstract
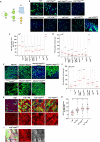
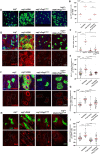
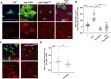
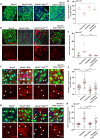
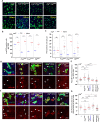
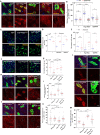
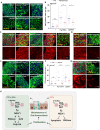
Tissue homeostasis requires a delicate balance between stem cell self-renewal, proliferation, and differentiation. Essential to this process is glycosylation, with both intra-and extra-cellular glycosylation being required for stem cell homeostasis. However, it remains unknown how intracellular glycosylation, O-GlcNAcylation, interfaces with cellular components of the extracellular glycosylation machinery, like the cytosolic N-glycanase NGLY1. In this study, we utilize the Drosophila gut and uncover a pathway in which O-GlcNAcylation cooperates with the NGLY1 homologue PNG1 to regulate proliferation in intestinal stem cells (ISCs) and apoptosis in differentiated enterocytes. Further, the CncC antioxidant signaling pathway and ENGase, an enzyme involved in the processing of free oligosaccharides in the cytosol, interact with O-GlcNAc and PNG1 through regulation of protein aggregates to contribute to gut maintenance. These findings reveal a complex coordinated regulation between O-GlcNAcylation and the cytosolic glycanase PNG1 critical to balancing proliferation and apoptosis to maintain gut homeostasis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Cytosolic N-GlcNAc proteins are formed by the action of endo-β-N-acetylglucosaminidase.Biochem Biophys Res Commun. 2020 Oct 1;530(4):719-724. doi: 10.1016/j.bbrc.2020.06.127. Epub 2020 Aug 8. Biochem Biophys Res Commun. 2020. PMID: 32782141 Free PMC article.
-
Drosophila EGFR pathway coordinates stem cell proliferation and gut remodeling following infection.BMC Biol. 2010 Dec 22;8:152. doi: 10.1186/1741-7007-8-152. BMC Biol. 2010. PMID: 21176204 Free PMC article.
-
O-GlcNAcylation: An Emerging Protein Modification Regulating the Hippo Pathway.Cancers (Basel). 2022 Jun 18;14(12):3013. doi: 10.3390/cancers14123013. Cancers (Basel). 2022. PMID: 35740678 Free PMC article. Review.
-
Enhancement of O-GlcNAcylation on Mitochondrial Proteins with 2-(4-Methoxyphenyl)ethyl-2-acetamido-2-deoxy-β-d-pyranoside, Contributes to the Mitochondrial Network, Cellular Bioenergetics and Stress Response in Neuronal Cells under Ischemic-like Conditions.Molecules. 2021 Sep 28;26(19):5883. doi: 10.3390/molecules26195883. Molecules. 2021. PMID: 34641427 Free PMC article.
-
[O-GlcNAc glycosylation and regulation of cell signaling].Med Sci (Paris). 2010 Aug-Sep;26(8-9):753-9. doi: 10.1051/medsci/2010268-9753. Med Sci (Paris). 2010. PMID: 20819714 Review. French.
Cited by
-
N-glycoproteomics reveals distinct glycosylation alterations in NGLY1-deficient patient-derived dermal fibroblasts.J Inherit Metab Dis. 2023 Jan;46(1):76-91. doi: 10.1002/jimd.12557. Epub 2022 Oct 4. J Inherit Metab Dis. 2023. PMID: 36102038 Free PMC article.
References
-
- Pham LV, Bryant JL, Mendez R, Chen J, Tamayo AT, Xu-Monette ZY, et al.. Targeting the hexosamine biosynthetic pathway and O-linked N-acetylglucosamine cycling for therapeutic and imaging capabilities in diffuse large B-cell lymphoma. Oncotarget. 2016;7(49):80599–611. Epub 2016/10/08. doi: 10.18632/oncotarget.12413 ; PubMed Central PMCID: PMC5348344. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

