Molecular Mechanism of Porcine Epidemic Diarrhea Virus Cell Tropism
- PMID: 35285698
- PMCID: PMC9040822
- DOI: 10.1128/mbio.03739-21
Molecular Mechanism of Porcine Epidemic Diarrhea Virus Cell Tropism
Abstract
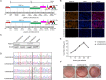
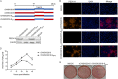
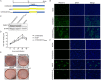
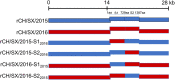
In the 21st century, several human and swine coronaviruses (CoVs) have emerged suddenly and caused great damage to people's lives and property. The porcine epidemic diarrhea virus (PEDV), leading to enormous economic losses to the pork industry and remains a large challenge. PEDV showed extensive cell tropism, and we cannot ignore the potential risk of cross-species transmission. However, the mechanism of adaptation and cell tropism of PEDV remains largely unknown and in vitro isolation of PEDV remains a huge challenge, which seriously impedes the development of vaccines. In this study, we confirmed that the spike (S) protein determines the adaptability of PEDV to monkey Vero cells and LLC-PK1 porcine cells, and isolated exchange of S1 and S2 subunits of adaptive strains did not make PEDV adapt to cells. Further, we found that the cellular adaptability of rCH/SX/2016-SHNXP depends on S1 and the first half of S2 (S3), and the 803L and 976H of the S2 subunit are critical for rCH/SX/2016-S1HNXP+S3HNXP adaptation to Vero cells. These findings highlight the decisive role of PEDV S protein in cell tropism and the potential role of coronaviruses S protein in cross-species transmissibility. Besides, our work also provides some different insight into finding PEDV receptors and developing PEDV and other coronaviruses vaccines. IMPORTANCE CoVs can spill from an animal reservoir into a naive host to cause diseases in humans or domestic animals. PEDV results in high mortality in piglets, which has caused immense economic losses in the pork industry. Virus isolation is the first step in studying viral pathogenesis and developing effective vaccines. However, the molecular mechanism of PEDV cell tropism is largely unknown, and isolation of endemic PEDV strains remains a major challenge. This study confirmed that the S gene is the decisive gene of PEDV adaptability to monkey Vero cells and porcine LLC-PK1 cells by the PEDV reverse genetics system. Isolated exchange of S1 and S2 of adaptive strains did not make PEDV adapt to cells, and the 803L and 976H of S2 subunit are critical for rCH/SX/2016-S1HNXP+S3HNXP adaptation to Vero cells. These results illustrate the decisive role of PEDV S protein in cell tropism and highlight the potential role of coronaviruses S protein in cross-species transmissibility. Besides, our finding also provides some unique insight into identifying PEDV functional receptors and has guiding significance for developing PEDV and other coronavirus vaccines.
Keywords: cellular tropism; coronavirus; porcine epidemic diarrhea virus; reverse genetic analysis; spike.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Development of a safe and broad-spectrum attenuated PEDV vaccine candidate by S2 subunit replacement.J Virol. 2024 Nov 19;98(11):e0042924. doi: 10.1128/jvi.00429-24. Epub 2024 Oct 15. J Virol. 2024. PMID: 39404450
-
Three Amino Acid Substitutions in the Spike Protein Enable the Coronavirus Porcine Epidemic Diarrhea Virus To Infect Vero Cells.Microbiol Spectr. 2023 Feb 14;11(1):e0387222. doi: 10.1128/spectrum.03872-22. Epub 2022 Dec 13. Microbiol Spectr. 2023. PMID: 36511700 Free PMC article.
-
Deletion of a 7-amino-acid region in the porcine epidemic diarrhea virus envelope protein induces higher type I and III interferon responses and results in attenuation in vivo.J Virol. 2023 Sep 28;97(9):e0084723. doi: 10.1128/jvi.00847-23. Epub 2023 Sep 8. J Virol. 2023. PMID: 37681956 Free PMC article.
-
Porcine epidemic diarrhea virus: Molecular mechanisms of attenuation and vaccines.Microb Pathog. 2020 Dec;149:104553. doi: 10.1016/j.micpath.2020.104553. Epub 2020 Oct 1. Microb Pathog. 2020. PMID: 33011361 Free PMC article. Review.
-
Porcine epidemic diarrhea virus (PEDV): An update on etiology, transmission, pathogenesis, and prevention and control.Virus Res. 2020 Sep;286:198045. doi: 10.1016/j.virusres.2020.198045. Epub 2020 Jun 2. Virus Res. 2020. PMID: 32502552 Free PMC article. Review.
Cited by
-
Research progress of porcine epidemic diarrhea virus S protein.Front Microbiol. 2024 May 30;15:1396894. doi: 10.3389/fmicb.2024.1396894. eCollection 2024. Front Microbiol. 2024. PMID: 38873162 Free PMC article. Review.
-
Porcine Epidemic Diarrhea: Insights and Progress on Vaccines.Vaccines (Basel). 2024 Feb 18;12(2):212. doi: 10.3390/vaccines12020212. Vaccines (Basel). 2024. PMID: 38400195 Free PMC article. Review.
-
Reverse Genetics Systems for Emerging and Re-Emerging Swine Coronaviruses and Applications.Viruses. 2023 Sep 26;15(10):2003. doi: 10.3390/v15102003. Viruses. 2023. PMID: 37896780 Free PMC article. Review.
-
A novel double antibody sandwich quantitative ELISA for detecting porcine epidemic diarrhea virus infection.Appl Microbiol Biotechnol. 2024 Oct 8;108(1):482. doi: 10.1007/s00253-024-13321-0. Appl Microbiol Biotechnol. 2024. PMID: 39377803 Free PMC article.
-
PEDV: Insights and Advances into Types, Function, Structure, and Receptor Recognition.Viruses. 2022 Aug 9;14(8):1744. doi: 10.3390/v14081744. Viruses. 2022. PMID: 36016366 Free PMC article. Review.
References
-
- Li W, Hulswit RJG, Kenney SP, Widjaja I, Jung K, Alhamo MA, van Dieren B, van Kuppeveld FJM, Saif LJ, Bosch BJ. 2018. Broad receptor engagement of an emerging global coronavirus may potentiate its diverse cross-species transmissibility. Proc Natl Acad Sci USA 115:E5135–E5143. doi:10.1073/pnas.1802879115. - DOI - PMC - PubMed
-
- Shi J, Wen Z, Zhong G, Yang H, Wang C, Huang B, Liu R, He X, Shuai L, Sun Z, Zhao Y, Liu P, Liang L, Cui P, Wang J, Zhang X, Guan Y, Tan W, Wu G, Chen H, Bu Z. 2020. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science 368:1016–1020. doi:10.1126/science.abb7015. - DOI - PMC - PubMed
-
- Edwards CE, Yount BL, Graham RL, Leist SR, Hou YJ, Dinnon KH, 3rd, Sims AC, Swanstrom J, Gully K, Scobey TD, Cooley MR, Currie CG, Randell SH, Baric RS. 2020. Swine acute diarrhea syndrome coronavirus replication in primary human cells reveals potential susceptibility to infection. Proc Natl Acad Sci USA 117:26915–26925. doi:10.1073/pnas.2001046117. - DOI - PMC - PubMed
-
- Lednicky JA, Tagliamonte MS, White SK, Elbadry MA, Alam MM, Stephenson CJ, Bonny TS, Loeb JC, Telisma T, Chavannes S, Ostrov DA, Mavian C, Beau De Rochars VM, Salemi M, Morris JG. Jr, 2021. Independent infections of porcine deltacoronavirus among Haitian children. Nature 600:133–137. doi:10.1038/s41586-021-04111-z. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- 2021JC-18/Science Foundation for Distinguished Young Scholars of Shaanxi Province
- CAAS-ASTIP-JBGS-20210602/Chinese Academy of Agricultural Science and Technology Innovation Project
- SKLVEB2020KFKT017/Open Project of the State Key Laboratory of Veterinary Etiological Biology
- Youth Innovation Team of Shaanxi Universities
- 293039/China Postdoctoral Science Foundation | Postdoctoral Research Foundation of China (China Postdoctoral Research Foundation)
LinkOut - more resources
Full Text Sources

