TSG-6 promotes Cancer Cell aggressiveness in a CD44-Dependent Manner and Reprograms Normal Fibroblasts to create a Pro-metastatic Microenvironment in Colorectal Cancer
- PMID: 35280699
- PMCID: PMC8898369
- DOI: 10.7150/ijbs.69178
TSG-6 promotes Cancer Cell aggressiveness in a CD44-Dependent Manner and Reprograms Normal Fibroblasts to create a Pro-metastatic Microenvironment in Colorectal Cancer
Abstract
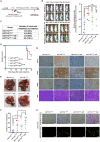
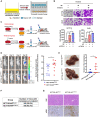
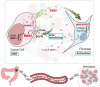
Tumor necrosis factor α stimulated gene 6 (TSG-6), a 30-KD secretory protein, plays an essential role in modulating inflammatory responses and extracellular matrix remodeling. However, little is known regarding the role of TSG-6 in human cancers. Here, we investigated the mechanism of action and the role of TSG-6 in colorectal cancer (CRC) metastasis. We found that TSG-6 was highly expressed in tumor tissues and was associated with poor prognosis and metastasis in CRC. Mechanistically, TSG-6 overexpression in CRC cells resulted in ERK activation and epithelial-mesenchymal transition by means of stabilizing CD44 and facilitating the CD44-EGFR complex formation on the cell membrane. Consequently, this resulted in the promotion of tumor migration and invasion both in vitro and in vivo. Notably, our data showed that CRC cells secreted TSG-6 could trigger a paracrine activation of JAK2-STAT3 signaling and reprogram normal fibroblasts into cancer-associated fibroblasts, which exhibited upregulation of pro-metastatic cytokines (CCL5 and MMP3) and higher movement ability. In animal models, the co-injection of cancer cells and TSG6-reprogrammed fibroblasts led to a significant increase in tumor metastasis. Our findings indicated that TSG-6 overexpression in CRC cells could promote cancer metastasis in both an autocrine and paracrine manner. Therefore, targeting TSG-6 might be a potential therapeutic strategy for the treatment of metastatic CRC.
Keywords: Colorectal Cancer; Fibroblast; Metastasis; TSG-6.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures

Similar articles
-
Colorectal cancer-associated fibroblasts promote metastasis by up-regulating LRG1 through stromal IL-6/STAT3 signaling.Cell Death Dis. 2021 Dec 20;13(1):16. doi: 10.1038/s41419-021-04461-6. Cell Death Dis. 2021. PMID: 34930899 Free PMC article.
-
Tumor necrosis factor receptor 2/AKT and ERK signaling pathways contribute to the switch from fibroblasts to CAFs by progranulin in microenvironment of colorectal cancer.Oncotarget. 2017 Apr 18;8(16):26323-26333. doi: 10.18632/oncotarget.15461. Oncotarget. 2017. PMID: 28412748 Free PMC article.
-
Primary tumors release ITGBL1-rich extracellular vesicles to promote distal metastatic tumor growth through fibroblast-niche formation.Nat Commun. 2020 Mar 5;11(1):1211. doi: 10.1038/s41467-020-14869-x. Nat Commun. 2020. PMID: 32139701 Free PMC article.
-
The Role of Adipocytes Recruited as Part of Tumor Microenvironment in Promoting Colorectal Cancer Metastases.Int J Mol Sci. 2024 Jul 30;25(15):8352. doi: 10.3390/ijms25158352. Int J Mol Sci. 2024. PMID: 39125923 Free PMC article. Review.
-
Cytokine- and chemokine-induced inflammatory colorectal tumor microenvironment: Emerging avenue for targeted therapy.Cancer Commun (Lond). 2022 Aug;42(8):689-715. doi: 10.1002/cac2.12295. Epub 2022 Jul 5. Cancer Commun (Lond). 2022. PMID: 35791509 Free PMC article. Review.
Cited by
-
Hyperplastic ovarian stromal cells express genes associated to tumor progression: a case study.BMC Vet Res. 2024 Sep 28;20(1):439. doi: 10.1186/s12917-024-04275-6. BMC Vet Res. 2024. PMID: 39342193 Free PMC article.
-
DKC1 aggravates gastric cancer cell migration and invasion through up-regulating the expression of TNFAIP6.Funct Integr Genomics. 2024 Feb 20;24(2):38. doi: 10.1007/s10142-024-01313-2. Funct Integr Genomics. 2024. PMID: 38376551 Free PMC article.
-
TNFAIP6 Promotes Gastric Carcinoma Cell Invasion via Upregulating PTX3 and Activating the Wnt/β-Catenin Signaling Pathway.Contrast Media Mol Imaging. 2022 Jul 6;2022:5697034. doi: 10.1155/2022/5697034. eCollection 2022. Contrast Media Mol Imaging. 2022. Retraction in: Contrast Media Mol Imaging. 2023 Jul 26;2023:9823836. doi: 10.1155/2023/9823836. PMID: 35854776 Free PMC article. Retracted.
-
Macrophage induces anti-cancer drug resistance in canine mammary gland tumor spheroid.Sci Rep. 2023 Jun 27;13(1):10394. doi: 10.1038/s41598-023-37311-w. Sci Rep. 2023. PMID: 37369757 Free PMC article.
-
Host Subcellular Organelles: Targets of Viral Manipulation.Int J Mol Sci. 2024 Jan 29;25(3):1638. doi: 10.3390/ijms25031638. Int J Mol Sci. 2024. PMID: 38338917 Free PMC article. Review.
References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–49. - PubMed
-
- Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC. et al. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70:145–64. - PubMed
-
- Thorsteinsson M, Jess P. The clinical significance of circulating tumor cells in non-metastatic colorectal cancer-a review. Eur J Surg Oncol. 2011;37:459–65. - PubMed
-
- Dienstmann R, Vermeulen L, Guinney J, Kopetz S, Tejpar S, Tabernero J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat Rev Cancer. 2017;17:268. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

