High glucose inhibits the osteogenic differentiation of periodontal ligament stem cells in periodontitis by activating endoplasmic reticulum stress
- PMID: 35280397
- PMCID: PMC8908123
- DOI: 10.21037/atm-22-6
High glucose inhibits the osteogenic differentiation of periodontal ligament stem cells in periodontitis by activating endoplasmic reticulum stress
Abstract
Background: Periodontitis is a highly prevalent dental disease which is associated with diabetes and is challenging to cure in diabetic patients. However, the mechanism of comorbid diabetes and periodontitis is still unclear. This study aimed to uncover the role of endoplasmic reticulum (ER) stress in high glucose-associated periodontitis.
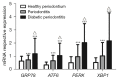
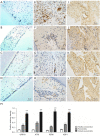
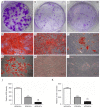
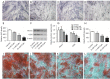
Methods: Periodontal tissues were obtained from diabetic patients with periodontitis, periodontitis patients without systemic disease, and healthy teeth. The expressions of ER stress-related factors GRP78, ATF6, PERK and XBP1 were detected by quantitative real-time polymerase chain reaction (qRT-PCR) and immumohistochemical staining. Periodontal ligament stem cells (PDLSCs) from three states of periodontal tissues were isolated and cultured as diabetic PDLSCs (dPDLSCs), inflamed PDLSCs (iPDLSCs) and healthy PDLSCs (hPDLSCs), and the cell stemness was assayed. Different concentrations (8, 11, and 25 mmol/L) of D-glucose were used on hPDLSCs to simulate high glucose microenvironment. The changes of osteogenic ability of PDLSCs were observed, and the expressions of ER stress-related factors in different time point (6, 12, 24, and 72 h) were detected. Finally, GRP78 shRNA lentivirus was used to block ER stress on PDLSCs in the 25 mmol/L D-glucose microenvironment, and the osteogenic ability of PDLSCs was observed.
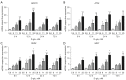
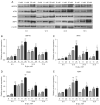
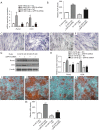
Results: The results showed that the expressions of GRP78, ATF6, PERK, and XBP1 were highest in the diabetic periodontitis group and lowest in the healthy periodontal tissue group (P<0.05). The clone formation, osteogenic and lipogenic differentiation abilities were lowest in dPDLSCs and highest in hPDLSCs. With the increase of glucose concentration, the osteogensis ability of PDLSCs decreased. After 6 hours of stimulation with D-glucose 25 mmol/L, the ER stress pathways in PDLSCs were effectively activated, and the peak value was reached at 12 hours. The decrease in the osteogensis ability of PDLSCs in a high glucose microenvironment reversed when ER stress was blocked.
Conclusions: The osteogenic differentiation ability of PDLSCs cells is inhibited in a high glucose microenvironment, and this effect is realized by ER stress activation. Blocking ER stress can partially restore the reduced osteogenic ability of PDLSCs. These results suggest that high glucose inhibits the osteogenic differentiation ability of PDLSCs by activating ER stress, which ultimately exacerbates periodontitis.
Keywords: Periodontitis; diabetes; endoplasmic reticulum stress; osteogenesis; periodontal ligament stem cells (PDLSCs).
2022 Annals of Translational Medicine. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-6/coif). The authors report that this work was supported by the National Natural Science Foundation of China (No. 81700987). The authors have no other conflicts of interest to declare.
Figures



Similar articles
-
Tumor Necrosis Factor-α Attenuates the Osteogenic Differentiation Capacity of Periodontal Ligament Stem Cells by Activating PERK Signaling.J Periodontol. 2016 Aug;87(8):e159-71. doi: 10.1902/jop.2016.150718. Epub 2016 Apr 18. J Periodontol. 2016. PMID: 27086613
-
Low-intensity pulsed ultrasound upregulates osteogenesis under inflammatory conditions in periodontal ligament stem cells through unfolded protein response.Stem Cell Res Ther. 2020 Jun 3;11(1):215. doi: 10.1186/s13287-020-01732-5. Stem Cell Res Ther. 2020. PMID: 32493507 Free PMC article.
-
Reuterin isolated from the probiotic Lactobacillus reuteri promotes periodontal tissue regeneration by inhibiting Cx43-mediated the intercellular transmission of endoplasmic reticulum stress.J Periodontal Res. 2024 Jun;59(3):552-564. doi: 10.1111/jre.13233. Epub 2024 Jan 9. J Periodontal Res. 2024. PMID: 38193526
-
Epigenetic regulation of osteogenic differentiation of periodontal ligament stem cells in periodontitis.Oral Dis. 2023 Oct;29(7):2529-2537. doi: 10.1111/odi.14491. Epub 2023 Jan 13. Oral Dis. 2023. PMID: 36582112 Review.
-
Overview of the main biological mechanisms linked to changes in periodontal ligament stem cells and the inflammatory microenvironment.J Zhejiang Univ Sci B. 2023 Apr 15;24(5):373-386. doi: 10.1631/jzus.B2200576. J Zhejiang Univ Sci B. 2023. PMID: 37190887 Free PMC article. Review.
Cited by
-
Ligament Alteration in Diabetes Mellitus.J Clin Med. 2022 Sep 27;11(19):5719. doi: 10.3390/jcm11195719. J Clin Med. 2022. PMID: 36233586 Free PMC article. Review.
-
Identification of Endoplasmic Reticulum Stress-Related Biomarkers of Periodontitis Based on Machine Learning: A Bioinformatics Analysis.Dis Markers. 2022 Aug 29;2022:8611755. doi: 10.1155/2022/8611755. eCollection 2022. Dis Markers. 2022. PMID: 36072904 Free PMC article. Clinical Trial.
-
IRE1α pathway: A potential bone metabolism mediator.Cell Prolif. 2024 Oct;57(10):e13654. doi: 10.1111/cpr.13654. Epub 2024 May 12. Cell Prolif. 2024. PMID: 38736291 Free PMC article. Review.
-
Endoplasmic reticulum stress: a novel targeted approach to repair bone defects by regulating osteogenesis and angiogenesis.J Transl Med. 2023 Jul 18;21(1):480. doi: 10.1186/s12967-023-04328-8. J Transl Med. 2023. PMID: 37464413 Free PMC article. Review.
-
Adipose-derived stem cell spheroid-laden microbial transglutaminase cross-linked gelatin hydrogel for treating diabetic periodontal wounds and craniofacial defects.Stem Cell Res Ther. 2023 Feb 3;14(1):20. doi: 10.1186/s13287-023-03238-2. Stem Cell Res Ther. 2023. PMID: 36737813 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous
