Secondary structure prediction for RNA sequences including N6-methyladenosine
- PMID: 35277476
- PMCID: PMC8917230
- DOI: 10.1038/s41467-022-28817-4
Secondary structure prediction for RNA sequences including N6-methyladenosine
Abstract
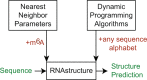
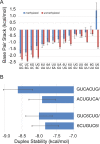
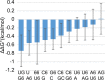
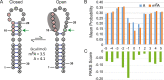
There is increasing interest in the roles of covalently modified nucleotides in RNA. There has been, however, an inability to account for modifications in secondary structure prediction because of a lack of software and thermodynamic parameters. We report the solution for these issues for N6-methyladenosine (m6A), allowing secondary structure prediction for an alphabet of A, C, G, U, and m6A. The RNAstructure software now works with user-defined nucleotide alphabets of any size. We also report a set of nearest neighbor parameters for helices and loops containing m6A, using experiments. Interestingly, N6-methylation decreases folding stability for adenosines in the middle of a helix, has little effect on folding stability for adenosines at the ends of helices, and increases folding stability for unpaired adenosines stacked on a helix. We demonstrate predictions for an N6-methylation-activated protein recognition site from MALAT1 and human transcriptome-wide effects of N6-methylation on the probability of adenosine being buried in a helix.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
A Test and Refinement of Folding Free Energy Nearest Neighbor Parameters for RNA Including N6-Methyladenosine.J Mol Biol. 2022 Sep 30;434(18):167632. doi: 10.1016/j.jmb.2022.167632. Epub 2022 May 16. J Mol Biol. 2022. PMID: 35588868 Free PMC article.
-
Secondary Structure Prediction of Single Sequences Using RNAstructure.Methods Mol Biol. 2016;1490:15-34. doi: 10.1007/978-1-4939-6433-8_2. Methods Mol Biol. 2016. PMID: 27665590
-
RNAstructure: software for RNA secondary structure prediction and analysis.BMC Bioinformatics. 2010 Mar 15;11:129. doi: 10.1186/1471-2105-11-129. BMC Bioinformatics. 2010. PMID: 20230624 Free PMC article.
-
The determination of RNA folding nearest neighbor parameters.Methods Mol Biol. 2014;1097:45-70. doi: 10.1007/978-1-62703-709-9_3. Methods Mol Biol. 2014. PMID: 24639154 Review.
-
How RNA folds.J Mol Biol. 1999 Oct 22;293(2):271-81. doi: 10.1006/jmbi.1999.3001. J Mol Biol. 1999. PMID: 10550208 Review.
Cited by
-
m6A readers, writers, erasers, and the m6A epitranscriptome in breast cancer.J Mol Endocrinol. 2022 Dec 21;70(2):e220110. doi: 10.1530/JME-22-0110. Print 2023 Feb 1. J Mol Endocrinol. 2022. PMID: 36367225 Free PMC article. Review.
-
How does precursor RNA structure influence RNA processing and gene expression?Biosci Rep. 2023 Mar 31;43(3):BSR20220149. doi: 10.1042/BSR20220149. Biosci Rep. 2023. PMID: 36689327 Free PMC article. Review.
-
Nearest neighbor rules for RNA helix folding thermodynamics: improved end effects.Nucleic Acids Res. 2022 May 20;50(9):5251-5262. doi: 10.1093/nar/gkac261. Nucleic Acids Res. 2022. PMID: 35524574 Free PMC article.
-
Nearest-neighbor parameters for the prediction of RNA duplex stability in diverse in vitro and cellular-like crowding conditions.Nucleic Acids Res. 2023 May 22;51(9):4101-4111. doi: 10.1093/nar/gkad020. Nucleic Acids Res. 2023. PMID: 36718808 Free PMC article.
-
A Test and Refinement of Folding Free Energy Nearest Neighbor Parameters for RNA Including N6-Methyladenosine.J Mol Biol. 2022 Sep 30;434(18):167632. doi: 10.1016/j.jmb.2022.167632. Epub 2022 May 16. J Mol Biol. 2022. PMID: 35588868 Free PMC article.
References
-
- Li X, Xiong X, Yi C. Epitranscriptome sequencing technologies: decoding RNA modifications. Nat. Methods. 2016;14:23–31. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

