Impaired p53-Mediated DNA Damage Response Contributes to Microcephaly in Nijmegen Breakage Syndrome Patient-Derived Cerebral Organoids
- PMID: 35269426
- PMCID: PMC8909307
- DOI: 10.3390/cells11050802
Impaired p53-Mediated DNA Damage Response Contributes to Microcephaly in Nijmegen Breakage Syndrome Patient-Derived Cerebral Organoids
Abstract
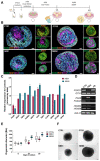
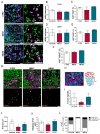
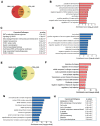
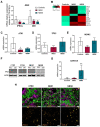
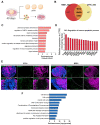
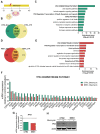
Nijmegen Breakage Syndrome (NBS) is a rare autosomal recessive genetic disorder caused by mutations within nibrin (NBN), a DNA damage repair protein. Hallmarks of NBS include chromosomal instability and clinical manifestations such as growth retardation, immunodeficiency, and progressive microcephaly. We employed induced pluripotent stem cell-derived cerebral organoids from two NBS patients to study the etiology of microcephaly. We show that NBS organoids carrying the homozygous 657del5 NBN mutation are significantly smaller with disrupted cyto-architecture. The organoids exhibit premature differentiation, and Neuronatin (NNAT) over-expression. Furthermore, pathways related to DNA damage response and cell cycle are differentially regulated compared to controls. After exposure to bleomycin, NBS organoids undergo delayed p53-mediated DNA damage response and aberrant trans-synaptic signaling, which ultimately leads to neuronal apoptosis. Our data provide insights into how mutations within NBN alters neurogenesis in NBS patients, thus providing a proof of concept that cerebral organoids are a valuable tool for studying DNA damage-related disorders.
Keywords: NBS; Neuronatin; cerebral organoids; disease modeling; iPSCs; microcephaly; p53.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Nijmegen Breakage Syndrome fibroblasts and iPSCs: cellular models for uncovering disease-associated signaling pathways and establishing a screening platform for anti-oxidants.Sci Rep. 2017 Aug 8;7(1):7516. doi: 10.1038/s41598-017-07905-2. Sci Rep. 2017. PMID: 28790359 Free PMC article.
-
Cleavage of the BRCT tandem domains of nibrin by the 657del5 mutation affects the DNA damage response less than the Arg215Trp mutation.IUBMB Life. 2012 Oct;64(10):853-61. doi: 10.1002/iub.1077. Epub 2012 Sep 3. IUBMB Life. 2012. PMID: 22941933
-
Nijmegen breakage syndrome (NBS).Orphanet J Rare Dis. 2012 Feb 28;7:13. doi: 10.1186/1750-1172-7-13. Orphanet J Rare Dis. 2012. PMID: 22373003 Free PMC article. Review.
-
Nijmegen breakage syndrome (NBS) with neurological abnormalities and without chromosomal instability.J Med Genet. 2006 Mar;43(3):218-24. doi: 10.1136/jmg.2005.035287. Epub 2005 Jul 20. J Med Genet. 2006. PMID: 16033915 Free PMC article.
-
Nijmegen breakage syndrome: case report and review of literature.Pan Afr Med J. 2020 Mar 20;35:85. doi: 10.11604/pamj.2020.35.85.14746. eCollection 2020. Pan Afr Med J. 2020. PMID: 32537088 Free PMC article. Review.
Cited by
-
Inhibition of the cGAS-STING pathway ameliorates the premature senescence hallmarks of Ataxia-Telangiectasia brain organoids.Aging Cell. 2021 Sep;20(9):e13468. doi: 10.1111/acel.13468. Epub 2021 Aug 30. Aging Cell. 2021. PMID: 34459078 Free PMC article.
-
Bilirubin-Induced Neurological Damage: Current and Emerging iPSC-Derived Brain Organoid Models.Cells. 2022 Aug 25;11(17):2647. doi: 10.3390/cells11172647. Cells. 2022. PMID: 36078055 Free PMC article. Review.
-
Cockayne Syndrome Patient iPSC-Derived Brain Organoids and Neurospheres Show Early Transcriptional Dysregulation of Biological Processes Associated with Brain Development and Metabolism.Cells. 2024 Mar 28;13(7):591. doi: 10.3390/cells13070591. Cells. 2024. PMID: 38607030 Free PMC article.
-
P53 independent pathogenic mechanisms contribute to BubR1 microcephaly.Front Cell Dev Biol. 2023 Oct 12;11:1282182. doi: 10.3389/fcell.2023.1282182. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37900274 Free PMC article.
-
The neurological and non-neurological roles of the primary microcephaly-associated protein ASPM.Front Neurosci. 2023 Aug 3;17:1242448. doi: 10.3389/fnins.2023.1242448. eCollection 2023. Front Neurosci. 2023. PMID: 37599996 Free PMC article. Review.
References
-
- Varon R., Demuth I., Chrzanowska K.H. Nijmegen Breakage Syndrome. University of Washington; Seattle, WA, USA: 1993. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

