How the ends signal the end: Regulation by E3 ubiquitin ligases recognizing protein termini
- PMID: 35247307
- PMCID: PMC9098119
- DOI: 10.1016/j.molcel.2022.02.004
How the ends signal the end: Regulation by E3 ubiquitin ligases recognizing protein termini
Abstract
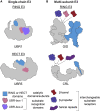
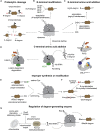
Specificity of eukaryotic protein degradation is determined by E3 ubiquitin ligases and their selective binding to protein motifs, termed "degrons," in substrates for ubiquitin-mediated proteolysis. From the discovery of the first substrate degron and the corresponding E3 to a flurry of recent studies enabled by modern systems and structural methods, it is clear that many regulatory pathways depend on E3s recognizing protein termini. Here, we review the structural basis for recognition of protein termini by E3s and how this recognition underlies biological regulation. Diverse E3s evolved to harness a substrate's N and/or C terminus (and often adjacent residues as well) in a sequence-specific manner. Regulation is achieved through selective activation of E3s and also through generation of degrons at ribosomes or by posttranslational means. Collectively, many E3 interactions with protein N and C termini enable intricate control of protein quality and responses to cellular signals.
Keywords: C-degron; E3 ligase; GID complex; N-degron; N-end rule; UBR; cullin-RING ligase; protein quality control; ubiquitin.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests B.A.S. is an honorary professor at the Technical University of Munich, Germany and adjunct faculty at St. Jude Children’s Research Hospital, Memphis, TN, USA. She is on the scientific advisory boards of Interline Therapeutics and BioTheryX and is a coinventor of intellectual property related to DCN1 small-molecule inhibitors licensed to Cinsano.
Figures




Similar articles
-
Structural basis for C-degron selectivity across KLHDCX family E3 ubiquitin ligases.Nat Commun. 2024 Nov 15;15(1):9899. doi: 10.1038/s41467-024-54126-z. Nat Commun. 2024. PMID: 39548056 Free PMC article.
-
Molecular basis for C-degron recognition by CRL2APPBP2 ubiquitin ligase.Proc Natl Acad Sci U S A. 2023 Oct 24;120(43):e2308870120. doi: 10.1073/pnas.2308870120. Epub 2023 Oct 16. Proc Natl Acad Sci U S A. 2023. PMID: 37844242 Free PMC article.
-
Protein quality control degron-containing substrates are differentially targeted in the cytoplasm and nucleus by ubiquitin ligases.Genetics. 2021 Mar 3;217(1):1-19. doi: 10.1093/genetics/iyaa031. Genetics. 2021. PMID: 33683364 Free PMC article.
-
Tying up loose ends: the N-degron and C-degron pathways of protein degradation.Biochem Soc Trans. 2020 Aug 28;48(4):1557-1567. doi: 10.1042/BST20191094. Biochem Soc Trans. 2020. PMID: 32627813 Free PMC article. Review.
-
Recognition of substrate degrons by E3 ubiquitin ligases and modulation by small-molecule mimicry strategies.Curr Opin Struct Biol. 2017 Jun;44:101-110. doi: 10.1016/j.sbi.2016.12.015. Epub 2017 Jan 25. Curr Opin Struct Biol. 2017. PMID: 28130986 Review.
Cited by
-
Recognition of the CCT5 di-Glu degron by CRL4DCAF12 is dependent on TRiC assembly.EMBO J. 2023 Feb 15;42(4):e112253. doi: 10.15252/embj.2022112253. Epub 2023 Jan 30. EMBO J. 2023. PMID: 36715408 Free PMC article.
-
Structure of the human UBR5 E3 ubiquitin ligase.Structure. 2023 May 4;31(5):541-552.e4. doi: 10.1016/j.str.2023.03.010. Epub 2023 Apr 10. Structure. 2023. PMID: 37040767 Free PMC article.
-
Mechanism and evolutionary origins of alanine-tail C-degron recognition by E3 ligases Pirh2 and CRL2-KLHDC10.Cell Rep. 2023 Sep 26;42(9):113100. doi: 10.1016/j.celrep.2023.113100. Epub 2023 Sep 6. Cell Rep. 2023. PMID: 37676773 Free PMC article.
-
Insights into the recognition mechanism in the UBR box of UBR4 for its specific substrates.Commun Biol. 2023 Nov 29;6(1):1214. doi: 10.1038/s42003-023-05602-7. Commun Biol. 2023. PMID: 38030679 Free PMC article.
-
The lowdown on breakdown: Open questions in plant proteolysis.Plant Cell. 2024 Sep 3;36(9):2931-2975. doi: 10.1093/plcell/koae193. Plant Cell. 2024. PMID: 38980154 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

