Interferons reshape the 3D conformation and accessibility of macrophage chromatin
- PMID: 35243225
- PMCID: PMC8857492
- DOI: 10.1016/j.isci.2022.103840
Interferons reshape the 3D conformation and accessibility of macrophage chromatin
Abstract
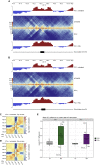
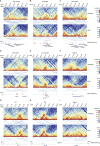
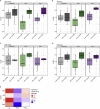
Engagement of macrophages in innate immune responses is directed by type I and type II interferons (IFN-I and IFN-γ, respectively). IFN triggers drastic changes in cellular transcriptomes, executed by JAK-STAT signal transduction and the transcriptional control of interferon-stimulated genes (ISG) by STAT transcription factors. Here, we study the immediate-early nuclear response to IFN-I and IFN-γ in murine macrophages. We show that the mechanism of gene control by both cytokines includes a rapid increase of DNA accessibility and rearrangement of the 3D chromatin contacts particularly between open chromatin of ISG loci. IFN-stimulated gene factor 3 (ISGF3), the major transcriptional regulator of ISG, controlled homeostatic and, most notably, induced-state DNA accessibility at a subset of ISG. Increases in DNA accessibility correlated with the appearance of activating histone marks at surrounding nucleosomes. Collectively our data emphasize changes in the three-dimensional nuclear space and epigenome as an important facet of transcriptional control by the IFN-induced JAK-STAT pathway.
Keywords: Cell biology; Epigenetics; Molecular biology; Molecular mechanism of gene regulation.
© 2022 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
A Positive Feedback Amplifier Circuit That Regulates Interferon (IFN)-Stimulated Gene Expression and Controls Type I and Type II IFN Responses.Front Immunol. 2018 May 28;9:1135. doi: 10.3389/fimmu.2018.01135. eCollection 2018. Front Immunol. 2018. PMID: 29892288 Free PMC article. Review.
-
Transcriptional and chromatin regulation in interferon and innate antiviral gene expression.Cytokine Growth Factor Rev. 2018 Dec;44:11-17. doi: 10.1016/j.cytogfr.2018.10.003. Epub 2018 Oct 22. Cytokine Growth Factor Rev. 2018. PMID: 30509403 Free PMC article. Review.
-
Two Interferon-Stimulated Response Elements Cooperatively Regulate Interferon-Stimulated Gene Expression in West Nile Virus-Infected IFNAR-/- Mouse Embryo Fibroblasts.J Virol. 2021 Oct 27;95(22):e0104021. doi: 10.1128/JVI.01040-21. Epub 2021 Sep 8. J Virol. 2021. PMID: 34495694 Free PMC article.
-
Interferon-gamma potentiates the antiviral activity and the expression of interferon-stimulated genes induced by interferon-alpha in U937 cells.J Interferon Res. 1992 Apr;12(2):87-94. doi: 10.1089/jir.1992.12.87. J Interferon Res. 1992. PMID: 1315834
-
Interferon regulatory factor-two restricts expression of interferon-stimulated genes to the endometrial stroma and glandular epithelium of the ovine uterus.Biol Reprod. 2001 Oct;65(4):1038-49. doi: 10.1095/biolreprod65.4.1038. Biol Reprod. 2001. PMID: 11566724
Cited by
-
Viral imitation is the sincerest form of epigenetic flattery.Mol Biol Cell. 2024 Oct 1;35(10):pe3. doi: 10.1091/mbc.E23-04-0147. Mol Biol Cell. 2024. PMID: 39302431 Free PMC article. Review.
-
Activation of human endogenous retroviruses and its physiological consequences.Nat Rev Mol Cell Biol. 2024 Mar;25(3):212-222. doi: 10.1038/s41580-023-00674-z. Epub 2023 Oct 23. Nat Rev Mol Cell Biol. 2024. PMID: 37872387 Review.
-
Mouse B2 SINE elements function as IFN-inducible enhancers.Elife. 2023 May 9;12:e82617. doi: 10.7554/eLife.82617. Elife. 2023. PMID: 37158599 Free PMC article.
-
The Culture Dish Surface Influences the Phenotype and Dissociation Strategy in Distinct Mouse Macrophage Populations.Front Immunol. 2022 Jul 6;13:920232. doi: 10.3389/fimmu.2022.920232. eCollection 2022. Front Immunol. 2022. PMID: 35874686 Free PMC article.
-
Merkel cell polyomavirus small tumor antigen contributes to immune evasion by interfering with type I interferon signaling.PLoS Pathog. 2024 Aug 7;20(8):e1012426. doi: 10.1371/journal.ppat.1012426. eCollection 2024 Aug. PLoS Pathog. 2024. PMID: 39110744 Free PMC article.
References
-
- Adelman K., Kennedy M.A., Nechaev S., Gilchrist D.A., Muse G.W., Chinenov Y., Rogatsky I. Immediate mediators of the inflammatory response are poised for gene activation through RNA polymerase II stalling. Proc. Natl. Acad. Sci. U S A. 2009;106:18207–18212. doi: 10.1073/pnas.0910177106. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

