Global analysis of miRNA-mRNA regulation pair in bladder cancer
- PMID: 35241117
- PMCID: PMC8896384
- DOI: 10.1186/s12957-022-02538-w
Global analysis of miRNA-mRNA regulation pair in bladder cancer
Abstract
Purpose: MicroRNA (miRNA) is a class of short non-coding RNA molecules that functions in RNA silencing and post-transcriptional regulation of gene expression. This study aims to identify critical miRNA-mRNA regulation pairs contributing to bladder cancer (BLCA) pathogenesis.
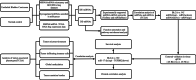
Patients and methods: MiRNA and mRNA microarray and RNA-sequencing datasets were downloaded from gene expression omnibus (GEO) and the cancer genome atlas (TCGA) databases. The tool of GEO2R and R packages were used to screen differential miRNAs (DE-miRNAs) and mRNAs (DE-mRNAs) and DAVID, DIANA, and Hiplot tools were used to perform gene enrichment analysis. The miRNA-mRNA regulation pair were screened from the experimentally validated miRNA-target interactions databases (miRTarbase and TarBase). Twenty-eight pairs of BLCA tissues were used to further verify the screened DE-miRNAs and DE-mRNAs by quantitative reverse transcription and polymerase chain reaction (qRT-PCR). The diagnostic value of the miRNA-mRNA regulation pairs was evaluated by receiver operating characteristic curve (ROC) and decision curve analysis (DCA). The correlation analysis between the selected miRNA-mRNAs regulation pair and clinical, survival and tumor-related phenotypes was performed in this study.
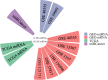
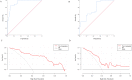
Results: After miRTarBase, the analysis of 2 miRNA datasets, 6 mRNA datasets, and TCGA-BLCA dataset, a total of 13 miRNAs (5 downregulated and 8 upregulated in BLCA tissues) and 181 mRNAs (72 upregulated and 109 downregulated in BLCA tissues) were screened out. The pairs of miR-17-5p (upregulated in BLCA tissues) and TGFBR2 (downregulated in BLCA tissues) were verified in the external validation cohort (28 BLCA vs. 28 NC) using qRT-PCR. Areas under the ROC curve of the miRNA-mRNA regulation pair panel were 0.929 (95% CI: 0.885-0.972, p < 0.0001) in TCGA-BLCA and 0.767 (95% CI: 0.643-0.891, p = 0.001) in the external validation. The DCA also showed that the miRNA-mRNA regulation pairs had an excellent diagnostic performance distinguishing BLCA from normal controls. Correlation analysis showed that miR-17-5p and TGFBR2 correlated with tumor immunity.
Conclusions: The research identified potential miRNA-mRNA regulation pairs, providing a new idea for exploring the genesis and development of BLCA.
Keywords: Bladder cancer; miRNA; miRNA-mRNA regulation pairs.
© 2022. The Author(s).
Conflict of interest statement
The author reports no conflicts of interest in this work.
Figures
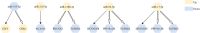


Similar articles
-
Identify miRNA-mRNA regulation pairs to explore potential pathogenesis of lung adenocarcinoma.Aging (Albany NY). 2022 Oct 19;14(20):8357-8373. doi: 10.18632/aging.204341. Epub 2022 Oct 19. Aging (Albany NY). 2022. PMID: 36260870 Free PMC article.
-
Construction of the miRNA-mRNA Regulatory Networks and Explore Their Role in the Development of Lung Squamous Cell Carcinoma.Front Mol Biosci. 2022 May 31;9:888020. doi: 10.3389/fmolb.2022.888020. eCollection 2022. Front Mol Biosci. 2022. PMID: 35712349 Free PMC article.
-
Analysis of aberrant miRNA-mRNA interaction networks in prostate cancer to conjecture its molecular mechanisms.Cancer Biomark. 2022;35(4):395-407. doi: 10.3233/CBM-220051. Cancer Biomark. 2022. PMID: 36373308
-
MicroRNAs: Their Role in Metastasis, Angiogenesis, and the Potential for Biomarker Utility in Bladder Carcinomas.Cancers (Basel). 2021 Feb 20;13(4):891. doi: 10.3390/cancers13040891. Cancers (Basel). 2021. PMID: 33672684 Free PMC article. Review.
-
Review of databases for experimentally validated human microRNA-mRNA interactions.Database (Oxford). 2023 Apr 25;2023:baad014. doi: 10.1093/database/baad014. Database (Oxford). 2023. PMID: 37098414 Free PMC article. Review.
Cited by
-
Construction of the underlying circRNA-miRNA-mRNA regulatory network and a new diagnostic model in ulcerative colitis by bioinformatics analysis.World J Clin Cases. 2024 Mar 26;12(9):1606-1621. doi: 10.12998/wjcc.v12.i9.1606. World J Clin Cases. 2024. PMID: 38576737 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

