Roles of transposable elements in the regulation of mammalian transcription
- PMID: 35228718
- PMCID: PMC10470143
- DOI: 10.1038/s41580-022-00457-y
Roles of transposable elements in the regulation of mammalian transcription
Abstract
Transposable elements (TEs) comprise about half of the mammalian genome. TEs often contain sequences capable of recruiting the host transcription machinery, which they use to express their own products and promote transposition. However, the regulatory sequences carried by TEs may affect host transcription long after the TEs have lost the ability to transpose. Recent advances in genome analysis and engineering have facilitated systematic interrogation of the regulatory activities of TEs. In this Review, we discuss diverse mechanisms by which TEs contribute to transcription regulation. Notably, TEs can donate enhancer and promoter sequences that influence the expression of host genes, modify 3D chromatin architecture and give rise to novel regulatory genes, including non-coding RNAs and transcription factors. We discuss how TEs spur regulatory evolution and facilitate the emergence of genetic novelties in mammalian physiology and development. By virtue of their repetitive and interspersed nature, TEs offer unique opportunities to dissect the effects of mutation and genomic context on the function and evolution of cis-regulatory elements. We argue that TE-centric studies hold the key to unlocking general principles of transcription regulation and evolution.
© 2022. Springer Nature Limited.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures



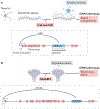
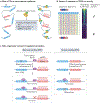
Similar articles
-
Transposable elements in mammalian chromatin organization.Nat Rev Genet. 2023 Oct;24(10):712-723. doi: 10.1038/s41576-023-00609-6. Epub 2023 Jun 7. Nat Rev Genet. 2023. PMID: 37286742 Review.
-
TFs for TEs: the transcription factor repertoire of mammalian transposable elements.Genes Dev. 2021 Jan 1;35(1-2):22-39. doi: 10.1101/gad.344473.120. Genes Dev. 2021. PMID: 33397727 Free PMC article. Review.
-
Transposable elements as a potent source of diverse cis-regulatory sequences in mammalian genomes.Philos Trans R Soc Lond B Biol Sci. 2020 Mar 30;375(1795):20190347. doi: 10.1098/rstb.2019.0347. Epub 2020 Feb 10. Philos Trans R Soc Lond B Biol Sci. 2020. PMID: 32075564 Free PMC article. Review.
-
Transposable Element Exaptation into Regulatory Regions Is Rare, Influenced by Evolutionary Age, and Subject to Pleiotropic Constraints.Mol Biol Evol. 2017 Nov 1;34(11):2856-2869. doi: 10.1093/molbev/msx219. Mol Biol Evol. 2017. PMID: 28961735 Free PMC article.
-
Jump-starting life: balancing transposable element co-option and genome integrity in the developing mammalian embryo.EMBO Rep. 2024 Apr;25(4):1721-1733. doi: 10.1038/s44319-024-00118-5. Epub 2024 Mar 25. EMBO Rep. 2024. PMID: 38528171 Free PMC article. Review.
Cited by
-
Transposon control as a checkpoint for tissue regeneration.Development. 2022 Nov 15;149(22):dev191957. doi: 10.1242/dev.191957. Epub 2022 Nov 28. Development. 2022. PMID: 36440631 Free PMC article.
-
Inferring Protein-DNA Binding Profiles at Interspersed Repeats Using HiChIP and PAtChER.Methods Mol Biol. 2023;2607:199-214. doi: 10.1007/978-1-0716-2883-6_11. Methods Mol Biol. 2023. PMID: 36449165
-
Ancestral genome reconstruction enhances transposable element annotation by identifying degenerate integrants.Cell Genom. 2024 Feb 14;4(2):100497. doi: 10.1016/j.xgen.2024.100497. Epub 2024 Jan 30. Cell Genom. 2024. PMID: 38295789 Free PMC article.
-
CRISPR-TE: a web-based tool to generate single guide RNAs targeting transposable elements.Mob DNA. 2024 Feb 1;15(1):3. doi: 10.1186/s13100-024-00313-0. Mob DNA. 2024. PMID: 38303094 Free PMC article.
-
Crosstalk between RNA m6A and DNA methylation regulates transposable element chromatin activation and cell fate in human pluripotent stem cells.Nat Genet. 2023 Aug;55(8):1324-1335. doi: 10.1038/s41588-023-01452-5. Epub 2023 Jul 20. Nat Genet. 2023. PMID: 37474847 Free PMC article.
References
-
- Fort V, Khelifi G & Hussein SMI Long non-coding RNAs and transposable elements: a functional relationship. Biochim. Biophys. Acta Mol. Cell Res 1868, 118837 (2021). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

