High-density genetic map and genome-wide association studies of aesthetic traits in Phalaenopsis orchids
- PMID: 35228611
- PMCID: PMC8885740
- DOI: 10.1038/s41598-022-07318-w
High-density genetic map and genome-wide association studies of aesthetic traits in Phalaenopsis orchids
Abstract
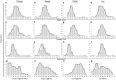
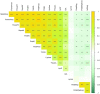
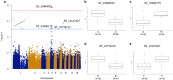
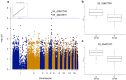
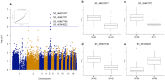
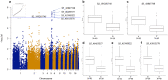
Phalaenopsis spp. represent the most popular orchids worldwide. Both P. equestris and P. aphrodite are the two important breeding parents with the whole genome sequence available. However, marker-trait association is rarely used for floral traits in Phalaenopsis breeding. Here, we analyzed markers associated with aesthetic traits of Phalaenopsis orchids by using genome-wide association study (GWAS) with the F1 population P. Intermedia of 117 progenies derived from the cross between P. aphrodite and P. equestris. A total of 113,517 single nucleotide polymorphisms (SNPs) were identified in P. Intermedia by using genotyping-by-sequencing with the combination of two different restriction enzyme pairs, Hinp1 I/Hae III and Apek I/Hae III. The size-related traits from flowers were negatively related to the color-related traits. The 1191 SNPs from Hinp1 I/ Hae III and 23 simple sequence repeats were used to establish a high-density genetic map of 19 homolog groups for P. equestris. In addition, 10 quantitative trait loci were highly associated with four color-related traits on chromosomes 2, 5 and 9. According to the sequence within the linkage disequilibrium regions, 35 candidate genes were identified and related to anthocyanin biosynthesis. In conclusion, we performed marker-assisted gene identification of aesthetic traits with GWAS in Phalaenopsis orchids.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
RNA-Seq SSRs of Moth Orchid and Screening for Molecular Markers across Genus Phalaenopsis (Orchidaceae).PLoS One. 2015 Nov 2;10(11):e0141761. doi: 10.1371/journal.pone.0141761. eCollection 2015. PLoS One. 2015. PMID: 26523377 Free PMC article.
-
The comparative chloroplast genomic analysis of photosynthetic orchids and developing DNA markers to distinguish Phalaenopsis orchids.Plant Sci. 2012 Jul;190:62-73. doi: 10.1016/j.plantsci.2012.04.001. Epub 2012 Apr 7. Plant Sci. 2012. PMID: 22608520
-
An overview of the Phalaenopsis orchid genome through BAC end sequence analysis.BMC Plant Biol. 2011 Jan 6;11:3. doi: 10.1186/1471-2229-11-3. BMC Plant Biol. 2011. PMID: 21208460 Free PMC article.
-
Genome-wide association study reveals novel quantitative trait loci and candidate genes of lint percentage in upland cotton based on the CottonSNP80K array.Theor Appl Genet. 2022 Jul;135(7):2279-2295. doi: 10.1007/s00122-022-04111-1. Epub 2022 May 16. Theor Appl Genet. 2022. PMID: 35570221 Review.
-
Breeding of ornamental orchids with focus on Phalaenopsis: current approaches, tools, and challenges for this century.Heredity (Edinb). 2024 Apr;132(4):163-178. doi: 10.1038/s41437-024-00671-8. Epub 2024 Feb 2. Heredity (Edinb). 2024. PMID: 38302667 Review.
Cited by
-
Genome-wide association analysis identified molecular markers and candidate genes for flower traits in Chinese orchid (Cymbidium sinense).Hortic Res. 2023 Oct 13;10(11):uhad206. doi: 10.1093/hr/uhad206. eCollection 2023 Nov. Hortic Res. 2023. PMID: 38046850 Free PMC article.
-
GWAS of adventitious root formation in roses identifies a putative phosphoinositide phosphatase (SAC9) for marker-assisted selection.PLoS One. 2023 Aug 18;18(8):e0287452. doi: 10.1371/journal.pone.0287452. eCollection 2023. PLoS One. 2023. PMID: 37595005 Free PMC article.
-
In-depth analysis of genomes and functional genomics of orchid using cutting-edge high-throughput sequencing.Front Plant Sci. 2022 Sep 23;13:1018029. doi: 10.3389/fpls.2022.1018029. eCollection 2022. Front Plant Sci. 2022. PMID: 36212315 Free PMC article. Review.
-
Advances and prospects of orchid research and industrialization.Hortic Res. 2022 Sep 28;9:uhac220. doi: 10.1093/hr/uhac220. eCollection 2022. Hortic Res. 2022. PMID: 36479582 Free PMC article.
-
Genome-Wide Association Study for Agro-Morphological Traits in Eggplant Core Collection.Plants (Basel). 2022 Oct 6;11(19):2627. doi: 10.3390/plants11192627. Plants (Basel). 2022. PMID: 36235493 Free PMC article.
References
-
- OrchidWiz. OrchidWiz encyclopedia version X4.2 June 2018 Database. Ames. IA (2018).
-
- Cai J, et al. The genome sequence of the orchid Phalaenopsis equestris. Nat. Genet. 2015;47:65–72. - PubMed
-
- Tsai WC, et al. OrchidBase 2.0: Comprehensive collection of Orchidaceae floral transcriptomes. Plant Cell Physiol. 2013;54:e7–e7. - PubMed
-
- Tsai W-C, Kuoh C-S, Chuang M-H, Chen W-H, Chen H-H. Four DEF-like MADS box genes displayed distinct floral morphogenetic roles in Phalaenopsis orchid. Plant Cell Physiol. 2004;45:831–844. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

