Trypanosoma Species in Small Nonflying Mammals in an Area With a Single Previous Chagas Disease Case
- PMID: 35223545
- PMCID: PMC8873152
- DOI: 10.3389/fcimb.2022.812708
Trypanosoma Species in Small Nonflying Mammals in an Area With a Single Previous Chagas Disease Case
Abstract
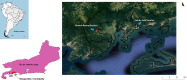
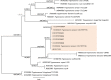
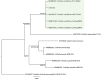
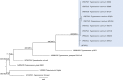
Trypanosomatids are hemoflagellate parasites that even though they have been increasingly studied, many aspects of their biology and taxonomy remain unknown. The aim of this study was to investigate the Trypanosoma sp. transmission cycle in nonflying small mammals in an area where a case of acute Chagas disease occurred in Mangaratiba municipality, Rio de Janeiro state. Three expeditions were conducted in the area: the first in 2012, soon after the human case, and two others in 2015. Sylvatic mammals were captured and submitted to blood collection for trypanosomatid parasitological and serological exams. Dogs from the surrounding areas where the sylvatic mammals were captured were also tested for T. cruzi infection. DNA samples were extracted from blood clots and positive hemocultures, submitted to polymerase chain reaction targeting SSU rDNA and gGAPDH genes, sequenced and phylogenetic analysed. Twenty-one wild mammals were captured in 2012, mainly rodents, and 17 mammals, mainly marsupials, were captured in the two expeditions conducted in 2015. Only four rodents demonstrated borderline serological T. cruzi test (IFAT), two in 2012 and two in 2015. Trypanosoma janseni was the main Trypanosoma species identified, and isolates were obtained solely from Didelphis aurita. In addition to biological differences, molecular differences are suggestive of genetic diversity in this flagellate species. Trypanosoma sp. DID was identified in blood clots from D. aurita in single and mixed infections with T. janseni. Concerning dogs, 12 presented mostly borderline serological titers for T. cruzi and no positive hemoculture. In blood clots from 11 dogs, T. cruzi DNA was detected and characterized as TcI (n = 9) or TcII (n = 2). Infections by Trypanosoma rangeli lineage E (n = 2) and, for the first time, Trypanosoma caninum, Trypanosoma dionisii, and Crithidia mellificae (n = 1 each) were also detected in dogs. We concluded that despite the low mammalian species richness and degraded environment, a high Trypanosoma species richness species was being transmitted with the predominance of T. janseni and not T. cruzi, as would be expected in a locality of an acute case of Chagas disease.
Keywords: Atlantic Forest; Trypanosoma cruzi clade; Trypanosomatidae; infection; mammalian host.
Copyright © 2022 Dario, Lisboa, Xavier, D’Andrea, Roque and Jansen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Trypanosomatid Richness Among Rats, Opossums, and Dogs in the Caatinga Biome, Northeast Brazil, a Former Endemic Area of Chagas Disease.Front Cell Infect Microbiol. 2022 Jun 20;12:851903. doi: 10.3389/fcimb.2022.851903. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35795183 Free PMC article.
-
Isolation and characterization of trypanosomatids, including Crithidia mellificae, in bats from the Atlantic Forest of Rio de Janeiro, Brazil.PLoS Negl Trop Dis. 2019 Jul 10;13(7):e0007527. doi: 10.1371/journal.pntd.0007527. eCollection 2019 Jul. PLoS Negl Trop Dis. 2019. PMID: 31291252 Free PMC article.
-
Chagas Immunochromatographic Rapid Test in the Serological Diagnosis of Trypanosoma cruzi Infection in Wild and Domestic Canids.Front Cell Infect Microbiol. 2022 Feb 22;12:835383. doi: 10.3389/fcimb.2022.835383. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35273924 Free PMC article.
-
Trypanosoma cruzi transmission in the wild and its most important reservoir hosts in Brazil.Parasit Vectors. 2018 Sep 6;11(1):502. doi: 10.1186/s13071-018-3067-2. Parasit Vectors. 2018. PMID: 30189896 Free PMC article. Review.
-
Landmarks of the Knowledge and Trypanosoma cruzi Biology in the Wild Environment.Front Cell Infect Microbiol. 2020 Feb 6;10:10. doi: 10.3389/fcimb.2020.00010. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32117794 Free PMC article. Review.
Cited by
-
First Molecular Identification of Trypanosomes and Absence of Babesia sp. DNA in Faeces of Non-Human Primates in the Ecuadorian Amazon.Pathogens. 2022 Dec 7;11(12):1490. doi: 10.3390/pathogens11121490. Pathogens. 2022. PMID: 36558823 Free PMC article.
-
Profile of natural Trypanosoma cruzi infection among dogs from rural areas of southern Espírito Santo, Brazil.Rev Soc Bras Med Trop. 2022 Dec 16;55:e0712-2021. doi: 10.1590/0037-8682-0712-2021. eCollection 2022. Rev Soc Bras Med Trop. 2022. PMID: 36542026 Free PMC article.
-
Kinetoplastid Species Maintained by a Small Mammal Community in the Pantanal Biome.Pathogens. 2022 Oct 19;11(10):1205. doi: 10.3390/pathogens11101205. Pathogens. 2022. PMID: 36297262 Free PMC article.
References
-
- Barbosa A. D., Mackie J. T., Stenner R., Gillett A., Irwin P., Ryan U. (2016). Trypanosoma Teixeirae: A New Species Belonging to the T. Cruzi Clade Causing Trypanosomosis in an Australian Little Red Flying Fox (Pteropus Scapulatus). Vet. Parasitol. 223, 214–221. doi: 10.1016/j.vetpar.2016.05.002 - DOI - PMC - PubMed
-
- Barros J. H., Almeida A. B., Figueiredo F. B., Sousa V. R., Fagundes A., Pinto A. G., et al. . (2012). Occurrence of Trypanosoma Caninum in Areas Overlapping With Leishmaniasis in Brazil: What is the Real Impact of Canine Leishmaniasis Control? Trans. R. Soc Trop. Med. Hyg. 106 (7), 419–423. doi: 10.1016/j.trstmh.2012.03.014 - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

