Tumor-specific T cells support chemokine-driven spatial organization of intratumoral immune microaggregates needed for long survival
- PMID: 35217577
- PMCID: PMC8883276
- DOI: 10.1136/jitc-2021-004346
Tumor-specific T cells support chemokine-driven spatial organization of intratumoral immune microaggregates needed for long survival
Abstract
Background: The composition of the tumor immune microenvironment (TIME) associated with good prognosis generally also predicts the success of immunotherapy, and both entail the presence of pre-existing tumor-specific T cells. Here, the blueprint of the TIME associated with such an ongoing tumor-specific T-cell response was dissected in a unique prospective oropharyngeal squamous cell carcinoma (OPSCC) cohort, in which tumor-specific tumor-infiltrating T cells were detected (immune responsiveness (IR+)) or not (lack of immune responsiveness (IR-)).
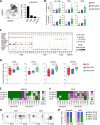
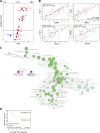
Methods: A comprehensive multimodal, high-dimensional strategy was applied to dissect the TIME of treatment-naive IR+ and IR- OPSCC tissue, including bulk RNA sequencing (NanoString), imaging mass cytometry (Hyperion) for phenotyping and spatial interaction analyses of immune cells, and combined single-cell gene expression profiling and T-cell receptor (TCR) sequencing (single-cell RNA sequencing (scRNAseq)) to characterize the transcriptional states of clonally expanded tumor-infiltrating T cells.
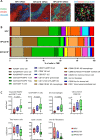
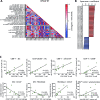
Results: IR+ patients had an excellent survival during >10 years follow-up. The tumors of IR+ patients expressed higher levels of genes strongly related to interferon gamma signaling, T-cell activation, TCR signaling, and mononuclear cell differentiation, as well as genes involved in several immune signaling pathways, than IR- patients. The top differently overexpressed genes included CXCL12 and LTB, involved in ectopic lymphoid structure development. Moreover, scRNAseq not only revealed that CD4+ T cells were the main producers of LTB but also identified a subset of clonally expanded CD8+ T cells, dominantly present in IR+ tumors, which secreted the T cell and dendritic cell (DC) attracting chemokine CCL4. Indeed, immune cell infiltration in IR+ tumors is stronger, highly coordinated, and has a distinct spatial phenotypical signature characterized by intratumoral microaggregates of CD8+CD103+ and CD4+ T cells with DCs. In contrast, the IR- TIME comprised spatial interactions between lymphocytes and various immunosuppressive myeloid cell populations. The impact of these chemokines on local immunity and clinical outcome was confirmed in an independent The Cancer Genome Atlas OPSCC cohort.
Conclusion: The production of lymphoid cell attracting and organizing chemokines by tumor-specific T cells in IR+ tumors constitutes a positive feedback loop to sustain the formation of the DC-T-cell microaggregates and identifies patients with excellent survival after standard therapy.
Keywords: immunotherapy; tumor microenvironment.
© Author(s) (or their employer(s)) 2022. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures



Similar articles
-
Intratumoral HPV16-Specific T Cells Constitute a Type I-Oriented Tumor Microenvironment to Improve Survival in HPV16-Driven Oropharyngeal Cancer.Clin Cancer Res. 2018 Feb 1;24(3):634-647. doi: 10.1158/1078-0432.CCR-17-2140. Epub 2017 Oct 10. Clin Cancer Res. 2018. PMID: 29018052
-
Heterodimeric IL-15 delays tumor growth and promotes intratumoral CTL and dendritic cell accumulation by a cytokine network involving XCL1, IFN-γ, CXCL9 and CXCL10.J Immunother Cancer. 2020 May;8(1):e000599. doi: 10.1136/jitc-2020-000599. J Immunother Cancer. 2020. PMID: 32461349 Free PMC article.
-
Tumor-infiltrating B cells affect the progression of oropharyngeal squamous cell carcinoma via cell-to-cell interactions with CD8+ T cells.J Immunother Cancer. 2019 Oct 17;7(1):261. doi: 10.1186/s40425-019-0726-6. J Immunother Cancer. 2019. PMID: 31623665 Free PMC article.
-
The Emerging Role of CD8+ Tissue Resident Memory T (TRM) Cells in Antitumor Immunity: A Unique Functional Contribution of the CD103 Integrin.Front Immunol. 2018 Aug 15;9:1904. doi: 10.3389/fimmu.2018.01904. eCollection 2018. Front Immunol. 2018. PMID: 30158938 Free PMC article. Review.
-
WNT/β-Catenin Signaling Pathway Regulating T Cell-Inflammation in the Tumor Microenvironment.Front Immunol. 2019 Sep 26;10:2293. doi: 10.3389/fimmu.2019.02293. eCollection 2019. Front Immunol. 2019. PMID: 31616443 Free PMC article. Review.
Cited by
-
CD4+ T cells produce IFN-I to license cDC1s for induction of cytotoxic T-cell activity in human tumors.Cell Mol Immunol. 2024 Apr;21(4):374-392. doi: 10.1038/s41423-024-01133-1. Epub 2024 Feb 21. Cell Mol Immunol. 2024. PMID: 38383773 Free PMC article.
-
Pivotal roles of tumor-draining lymph nodes in the abscopal effects from combined immunotherapy and radiotherapy.Cancer Commun (Lond). 2022 Oct;42(10):971-986. doi: 10.1002/cac2.12348. Epub 2022 Aug 13. Cancer Commun (Lond). 2022. PMID: 35962977 Free PMC article.
-
Single-cell high-dimensional imaging mass cytometry: one step beyond in oncology.Semin Immunopathol. 2023 Jan;45(1):17-28. doi: 10.1007/s00281-022-00978-w. Epub 2023 Jan 4. Semin Immunopathol. 2023. PMID: 36598557 Free PMC article. Review.
-
Monocyte infiltration is an independent positive prognostic biomarker in vulvar squamous cell carcinoma.Cancer Immunol Immunother. 2024 Jul 2;73(9):166. doi: 10.1007/s00262-024-03755-w. Cancer Immunol Immunother. 2024. PMID: 38954042 Free PMC article.
-
Transferrin receptor-targeted immunostimulant for photodynamic immunotherapy against metastatic tumors through β-catenin/CREB interruption.Acta Pharm Sin B. 2024 Sep;14(9):4118-4133. doi: 10.1016/j.apsb.2024.05.030. Epub 2024 Jun 3. Acta Pharm Sin B. 2024. PMID: 39309507 Free PMC article.
References
-
- Burtness B, Harrington KJ, Greil R, et al. . Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): a randomised, open-label, phase 3 study. Lancet 2019;394:1915–28. 10.1016/S0140-6736(19)32591-7 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
