Genome-Wide Identification and Gene Expression Analysis of the OTU DUB Family in Oryza sativa
- PMID: 35215984
- PMCID: PMC8878984
- DOI: 10.3390/v14020392
Genome-Wide Identification and Gene Expression Analysis of the OTU DUB Family in Oryza sativa
Abstract
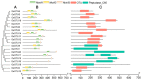
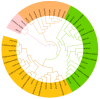
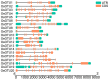
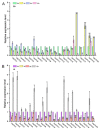
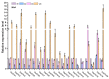
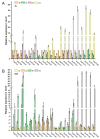
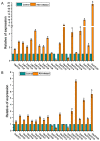
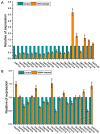
Ovarian tumor domain (OTU)-containing deubiquitinating enzymes (DUBs) are an essential DUB to maintain protein stability in plants and play important roles in plant growth development and stress response. However, there is little genome-wide identification and analysis of the OTU gene family in rice. In this study, we identified 20 genes of the OTU family in rice genome, which were classified into four groups based on the phylogenetic analysis. Their gene structures, conserved motifs and domains, chromosomal distribution, and cis elements in promoters were further studied. In addition, OTU gene expression patterns in response to plant hormone treatments, including SA, MeJA, NAA, BL, and ABA, were investigated by RT-qPCR analysis. The results showed that the expression profile of OsOTU genes exhibited plant hormone-specific expression. Expression levels of most of the rice OTU genes were significantly changed in response to rice stripe virus (RSV), rice black-streaked dwarf virus (RBSDV), Southern rice black-streaked dwarf virus (SRBSDV), and Rice stripe mosaic virus (RSMV). These results suggest that the rice OTU genes are involved in diverse hormone signaling pathways and in varied responses to virus infection, providing new insights for further functional study of OsOTU genes.
Keywords: OTU DUB; genome-wide; plant hormone treatment; rice; virus infection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Rice black-streaked dwarf virus P10 acts as either a synergistic or antagonistic determinant during superinfection with related or unrelated virus.Mol Plant Pathol. 2019 May;20(5):641-655. doi: 10.1111/mpp.12782. Epub 2019 Feb 14. Mol Plant Pathol. 2019. PMID: 30623552 Free PMC article.
-
Genome-Wide Analysis of the RAV Transcription Factor Genes in Rice Reveals Their Response Patterns to Hormones and Virus Infection.Viruses. 2021 Apr 25;13(5):752. doi: 10.3390/v13050752. Viruses. 2021. PMID: 33922971 Free PMC article.
-
Transcriptional changes of rice in response to rice black-streaked dwarf virus.Gene. 2017 Sep 10;628:38-47. doi: 10.1016/j.gene.2017.07.015. Epub 2017 Jul 9. Gene. 2017. PMID: 28700950
-
A Review of Vector-Borne Rice Viruses.Viruses. 2022 Oct 14;14(10):2258. doi: 10.3390/v14102258. Viruses. 2022. PMID: 36298813 Free PMC article. Review.
-
Exploring the shared pathogenic strategies of independently evolved effectors across distinct plant viruses.Trends Microbiol. 2024 Oct;32(10):1021-1033. doi: 10.1016/j.tim.2024.03.001. Epub 2024 Mar 22. Trends Microbiol. 2024. PMID: 38521726 Review.
Cited by
-
The Arabidopsis Deubiquitylase OTU5 Suppresses Flowering by Histone Modification-Mediated Activation of the Major Flowering Repressors FLC, MAF4, and MAF5.Int J Mol Sci. 2023 Mar 24;24(7):6176. doi: 10.3390/ijms24076176. Int J Mol Sci. 2023. PMID: 37047144 Free PMC article.
-
Plant deubiquitinases: from structure and activity to biological functions.Plant Cell Rep. 2023 Mar;42(3):469-486. doi: 10.1007/s00299-022-02962-y. Epub 2022 Dec 26. Plant Cell Rep. 2023. PMID: 36567335 Review.
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

