Orientin, a Bio-Flavonoid from Trigonella hamosa L., Regulates COX-2/PGE-2 in A549 Cell Lines via miR-26b and miR-146a
- PMID: 35215267
- PMCID: PMC8876523
- DOI: 10.3390/ph15020154
Orientin, a Bio-Flavonoid from Trigonella hamosa L., Regulates COX-2/PGE-2 in A549 Cell Lines via miR-26b and miR-146a
Abstract
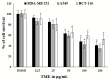
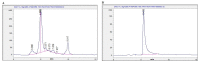
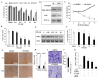
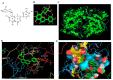
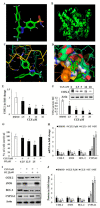
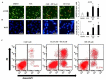
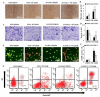
Cancer is a severe health condition and considered one of the major healthcare issues and is in need of innovative strategy for a cure. The current study aimed to investigate the chemical profile of Trigonella hamosa L. and a potential molecular approach to explain its regulation in cancer progression through an inflammatory mediator (COX-2) in A549 non-small lung cancer cell lines via in silico, mechanistic and molecular aspects. T. hamosa was extracted and then subjected to a CCK-8 cell viability assay in different cancer cell lines including MDA-MB-231, A549 and HCT-116. Total extract was subjected to several chromatographic techniques to yield orientin (OT); the structure was elucidated by inspection of NMR spectroscopic data. To achieve anticancer effects of OT, a cell viability assay using a CCK-8 kit, immunoprecipitation by Western blot, cell migration using a wound healing assay, cell invasion using a Matrigel-Transwell assay, apoptosis by AO/EB dual staining, flow cytometric analysis and DAPI staining, a silenced COX-2 model to determine PGE-2 production and real-time PCR and Western blot of BCL-2, CYP-1A1, iNOS and COX-2 markers were carried out. The results demonstrated that OT decreased the cell proliferation and controlled cell migration and invasive properties. OT destabilized the COX-2 mRNA and downregulated its expression in A549 cell lines. Virtual binding showed interaction (binding energy -10.43) between OT and COX-2 protein compared to the selective COX-2 inhibitor celecoxib (CLX) (binding energy -9.4). The OT-CLX combination showed a superior anticancer effect. The synergistic effect of OT-CLX combination was noticed in controlling the migration and invasion of A549 cell lines. OT-CLX downregulated the expression of BCL-2, iNOS and COX-2 and activated the proapoptotic gene CYP-1A1. OT mitigated the COX-2 expression via upregulation of miR-26b and miR-146a. Interestingly, COX-2-silenced transfected A549 cells exhibited reduced expression of miR-26b and miR-146a. The findings confirmed the direct interaction of OT with COX-2 protein. PGE-2 expression was quantified in both naïve and COX-2-silenced A549 cells. OT downregulated the release of PGE-2 in both tested conditions. These results confirmed the regulatory effect of OT on A549 cell growth in a COX-2-dependent manner. OT activated apoptosis via activation of CYP-1A1 expression in an independent manner. These results revealed that the OT-CLX combination could serve as a potential synergistic treatment for effective inflammatory-mediated anticancer strategies.
Keywords: A549; COX-2 inhibitor; Trigonella hamosa L.; lung cancer; miRNA; migration; orientin.
Conflict of interest statement
The authors declare that there are no conflict of interest.
Figures

Similar articles
-
MiR-26b suppresses tumor cell proliferation, migration and invasion by directly targeting COX-2 in lung cancer.Eur Rev Med Pharmacol Sci. 2015 Dec;19(24):4728-37. Eur Rev Med Pharmacol Sci. 2015. PMID: 26744864
-
lncRNA SOX2-OT regulates laryngeal cancer cell proliferation, migration and invasion and induces apoptosis by suppressing miR-654.Exp Ther Med. 2020 May;19(5):3316-3324. doi: 10.3892/etm.2020.8577. Epub 2020 Mar 6. Exp Ther Med. 2020. PMID: 32266028 Free PMC article.
-
[MiR-26b inhibits proliferation, invasion, and migration of glioma by targeting cyclooxygenase-2].Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2017 Feb 28;42(2):139-146. doi: 10.11817/j.issn.1672-7347.2017.02.004. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2017. PMID: 28255114 Chinese.
-
MicroRNA-126 Targeting PIK3R2 Inhibits NSCLC A549 Cell Proliferation, Migration, and Invasion by Regulation of PTEN/PI3K/AKT Pathway.Clin Lung Cancer. 2016 Sep;17(5):e65-e75. doi: 10.1016/j.cllc.2016.03.012. Epub 2016 Apr 6. Clin Lung Cancer. 2016. PMID: 27236384
-
Effect and mechanism of miR-146a on malignant biological behaviors of lung adenocarcinoma cell line.Oncol Lett. 2020 Jun;19(6):3643-3652. doi: 10.3892/ol.2020.11474. Epub 2020 Mar 23. Oncol Lett. 2020. PMID: 32382320 Free PMC article.
Cited by
-
Capric Acid Behaves Agonistic Effect on Calcitriol to Control Inflammatory Mediators in Colon Cancer Cells.Molecules. 2022 Oct 6;27(19):6624. doi: 10.3390/molecules27196624. Molecules. 2022. PMID: 36235161 Free PMC article.
-
A comprehensive account on ethnobotany, phytochemistry and pharmacological insights of genus Celtis.Heliyon. 2024 Apr 25;10(9):e29707. doi: 10.1016/j.heliyon.2024.e29707. eCollection 2024 May 15. Heliyon. 2024. PMID: 38726115 Free PMC article. Review.
-
Mechanistic Insights into the Ameliorative Effect of Cichoriin on Diabetic Rats-Assisted with an In Silico Approach.Molecules. 2022 Oct 24;27(21):7192. doi: 10.3390/molecules27217192. Molecules. 2022. PMID: 36364019 Free PMC article.
-
LC-TOF-MS/MS and GC-MS based phytochemical profiling and evaluation of wound healing activity of Oroxylum Indicum (L.) Kurz (Beka).Front Pharmacol. 2022 Nov 22;13:1050453. doi: 10.3389/fphar.2022.1050453. eCollection 2022. Front Pharmacol. 2022. PMID: 36483735 Free PMC article.
-
Polyphenols as Lung Cancer Chemopreventive Agents by Targeting microRNAs.Molecules. 2022 Sep 11;27(18):5903. doi: 10.3390/molecules27185903. Molecules. 2022. PMID: 36144639 Free PMC article. Review.
References
-
- Khalil H.E., Mohamed M.E., Morsy M.A., Kandeel M. Flavonoid and phenolic compounds from Carissa macrocarpa: Molecular docking and cytotoxicity studies. Pharmacogn. Mag. 2018;14:304. doi: 10.4103/pm.pm_104_18. - DOI
-
- Migahid A. Flora of Saudi Arabia. King Saud University Press; Riyadh, Saudi Arabia: 1987.
-
- Youssef S. Medicinal and non-medicinal uses of some plants found in the middle region of Saudi Arabia. J. Med. Plant Res. 2013;7:2501–2517.
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

